Michael Jendrusch
@mjendrusch.bsky.social
810 followers
310 following
33 posts
(he/him) Former PhD student / Postdoc @ Korbel group, EMBL.
Protein ML person, mathematics & science enthusiast.
developer of salad
preprint: https://www.biorxiv.org/content/10.1101/2025.01.31.635780v1
code: https://github.com/mjendrusch/salad
Posts
Media
Videos
Starter Packs
Pinned
Reposted by Michael Jendrusch
Reposted by Michael Jendrusch
Reposted by Michael Jendrusch
Reposted by Michael Jendrusch
Reposted by Michael Jendrusch
Reposted by Michael Jendrusch
Reposted by Michael Jendrusch
Reposted by Michael Jendrusch
Reposted by Michael Jendrusch
Reposted by Michael Jendrusch
Reposted by Michael Jendrusch
Reposted by Michael Jendrusch
Yo Akiyama
@yoakiyama.bsky.social
· Aug 5

Scaling down protein language modeling with MSA Pairformer
Recent efforts in protein language modeling have focused on scaling single-sequence models and their training data, requiring vast compute resources that limit accessibility. Although models that use ...
biorxiv.org
Reposted by Michael Jendrusch
Jan Mathony
@jmathony.bsky.social
· Aug 4

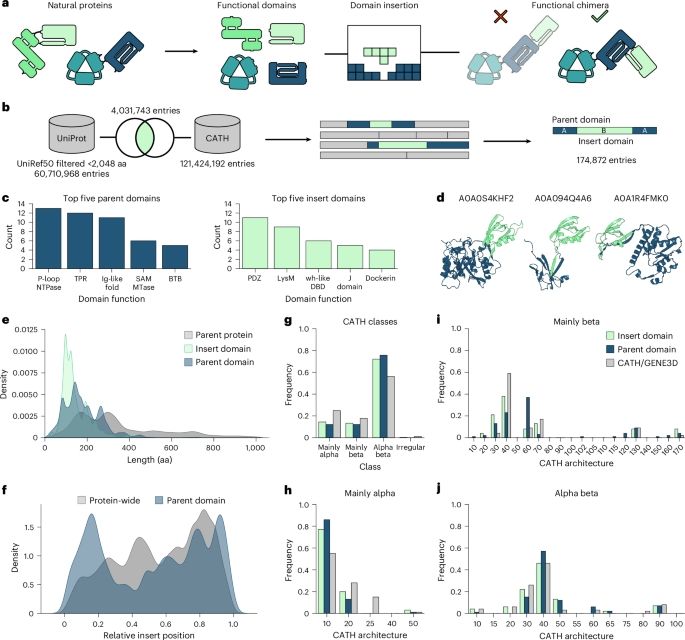
Rational engineering of allosteric protein switches by in silico prediction of domain insertion sites
Nature Methods - ProDomino is a machine leaning-based method, trained on a semisynthetic domain insertion dataset, to guide the engineering of protein domain recombination.
rdcu.be
Reposted by Michael Jendrusch