Maria Letizia Di Martino
@mldm.bsky.social
370 followers
620 following
16 posts
Senior Researcher at Uppsala University | Host-pathogen interactions | Intestinal organoids | Virulence gene regulation | Shigella and Salmonella | Functional genomics | Affiliated to the SellinLab @sellinlab.bsky.social
Posts
Media
Videos
Starter Packs
Reposted by Maria Letizia Di Martino
SellinLab
@sellinlab.bsky.social
· Aug 24

Mast Cell Phenotypic Heterogeneity Impacts the Interplay with Pathogenic Salmonella Typhimurium Bacteria
Mast cell phenotypic features influence anti-bacterial response. Connective tissue-like mast cells sense Salmonella through combined Toll-like-receptor (TLR) signaling and recognition of Type-III-sec....
onlinelibrary.wiley.com
Reposted by Maria Letizia Di Martino
SellinLab
@sellinlab.bsky.social
· Aug 5

Matrix-M adjuvant triggers inflammasome activation and enables antigen cross-presentation through induction of lysosomal membrane permeabilization - npj Vaccines
npj Vaccines - Matrix-M adjuvant triggers inflammasome activation and enables antigen cross-presentation through induction of lysosomal membrane permeabilization
www.nature.com
Reposted by Maria Letizia Di Martino
Nature Genetics
@natgenet.nature.com
· Jun 23

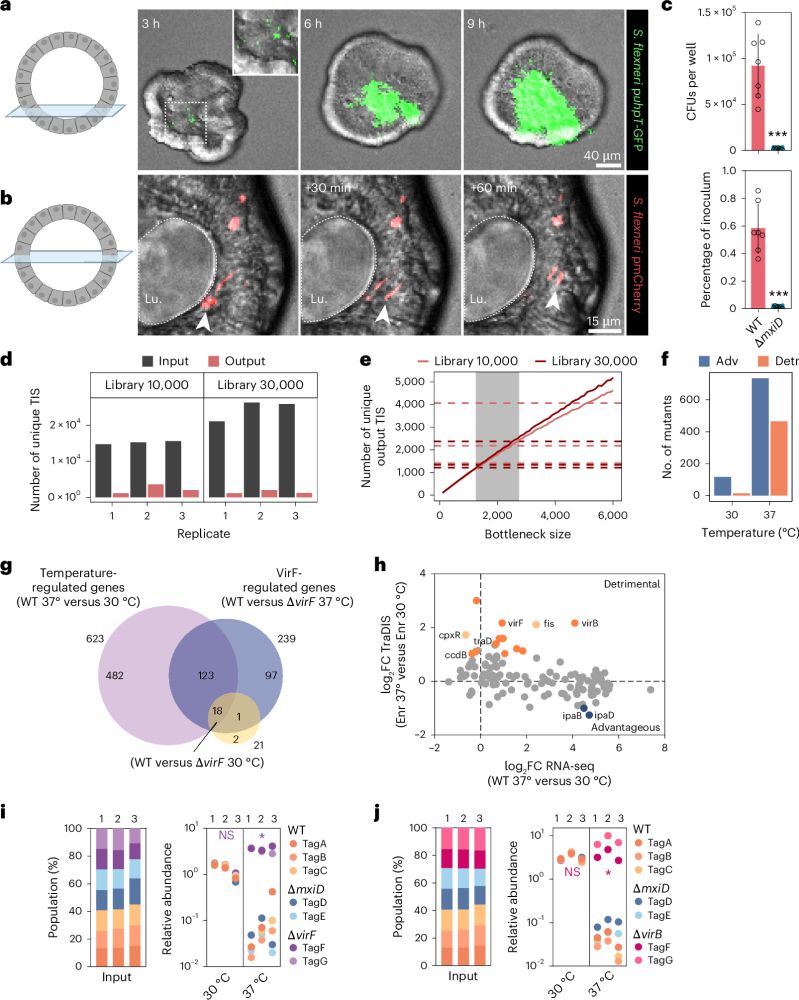
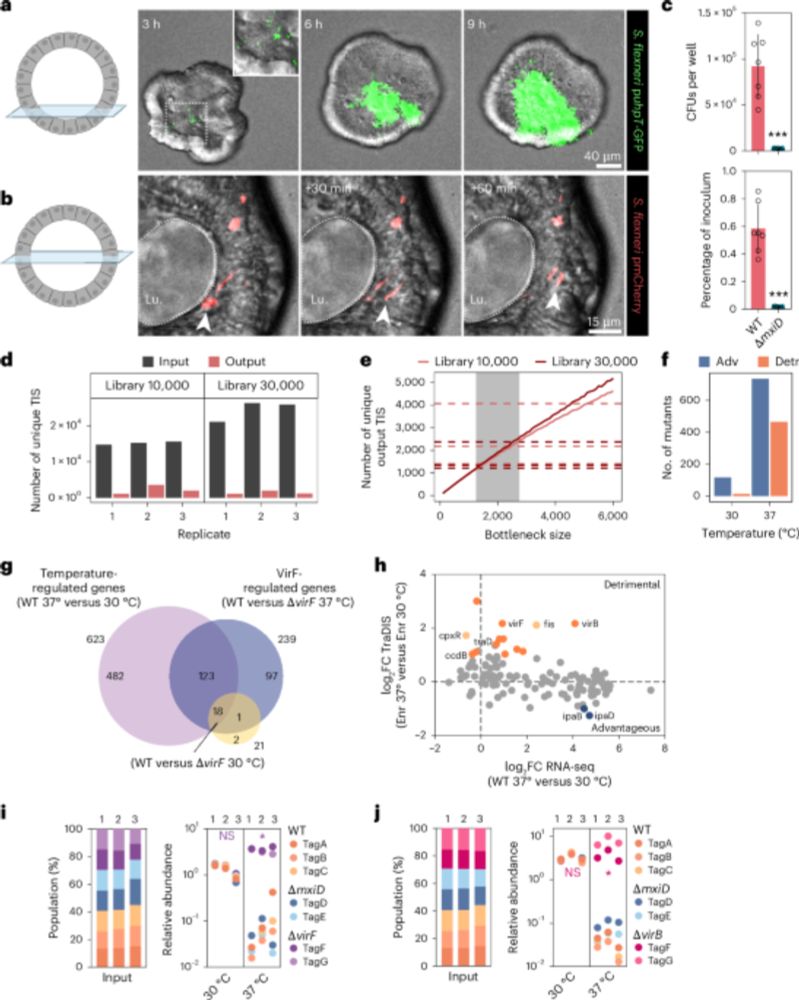
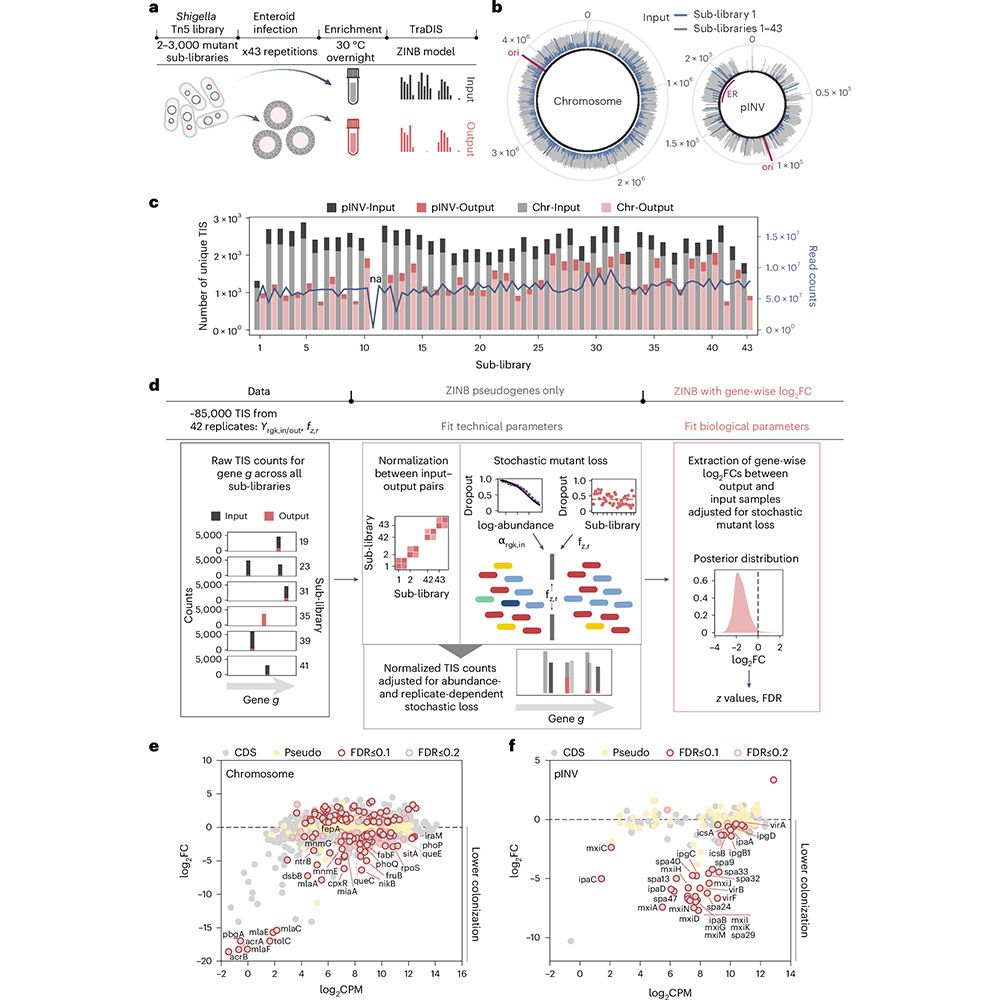
A scalable gut epithelial organoid model reveals the genome-wide colonization landscape of a human-adapted pathogen - Nature Genetics
A genome-wide screen using human gut epithelial organoids combined with transposon-directed insertion sequencing identifies over 100 Shigella flexneri genes required for epithelial colonization.
www.nature.com
Reposted by Maria Letizia Di Martino
Reposted by Maria Letizia Di Martino
Reposted by Maria Letizia Di Martino
SellinLab
@sellinlab.bsky.social
· Jun 13
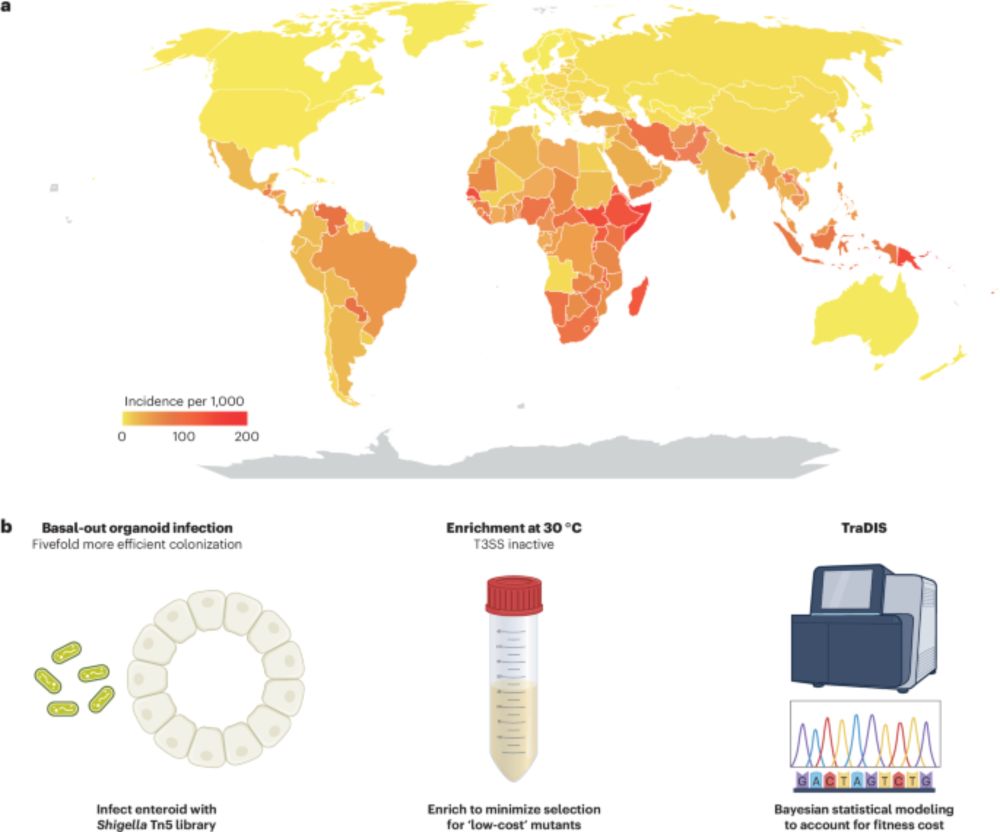
Organoid models advance dysentery control - Nature Genetics
Shigella is a major human pathogen with a stark lack of pipelines linking genome content to clinical pathogenesis. An innovative study using large-scale organoid models, combined with genome-wide muta...
www.nature.com
Reposted by Maria Letizia Di Martino
SellinLab
@sellinlab.bsky.social
· Jun 12
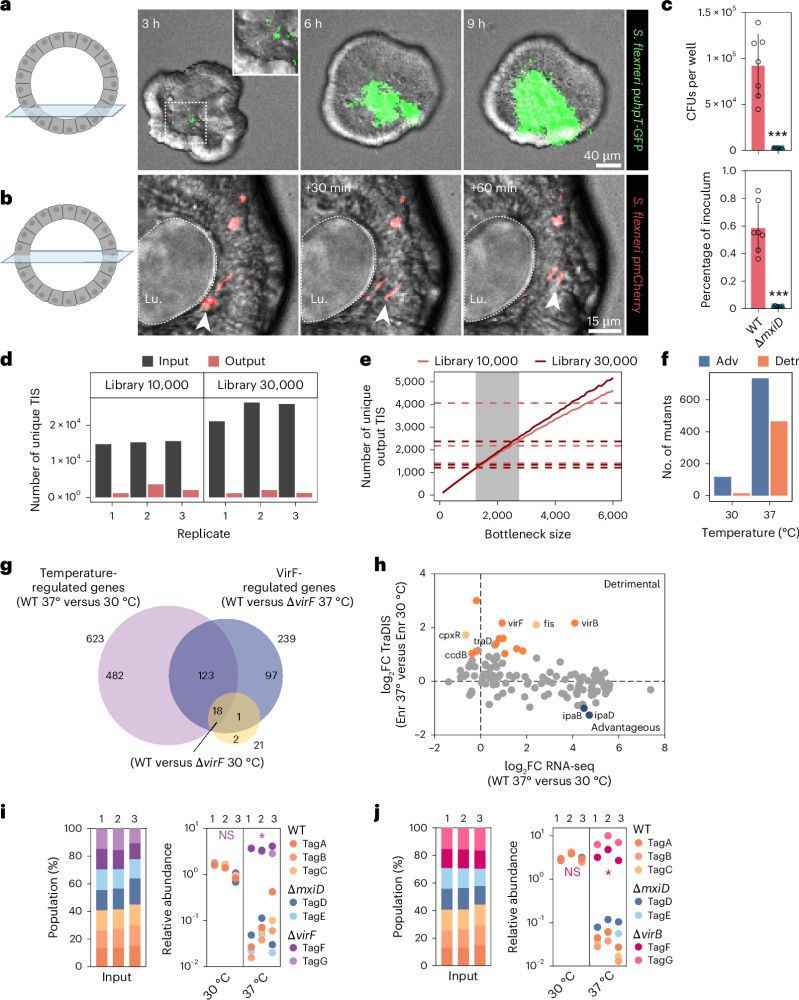
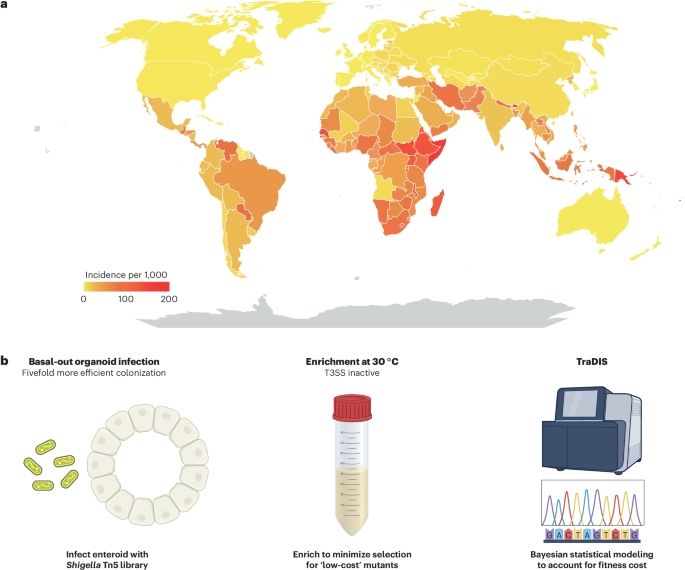
A scalable gut epithelial organoid model reveals the genome-wide colonization landscape of a human-adapted pathogen
Nature Genetics - A genome-wide screen using human gut epithelial organoids combined with transposon-directed insertion sequencing identifies over 100 Shigella flexneri genes required for...
rdcu.be
Reposted by Maria Letizia Di Martino
Lars Barquist
@lbarquist.bsky.social
· Jun 12
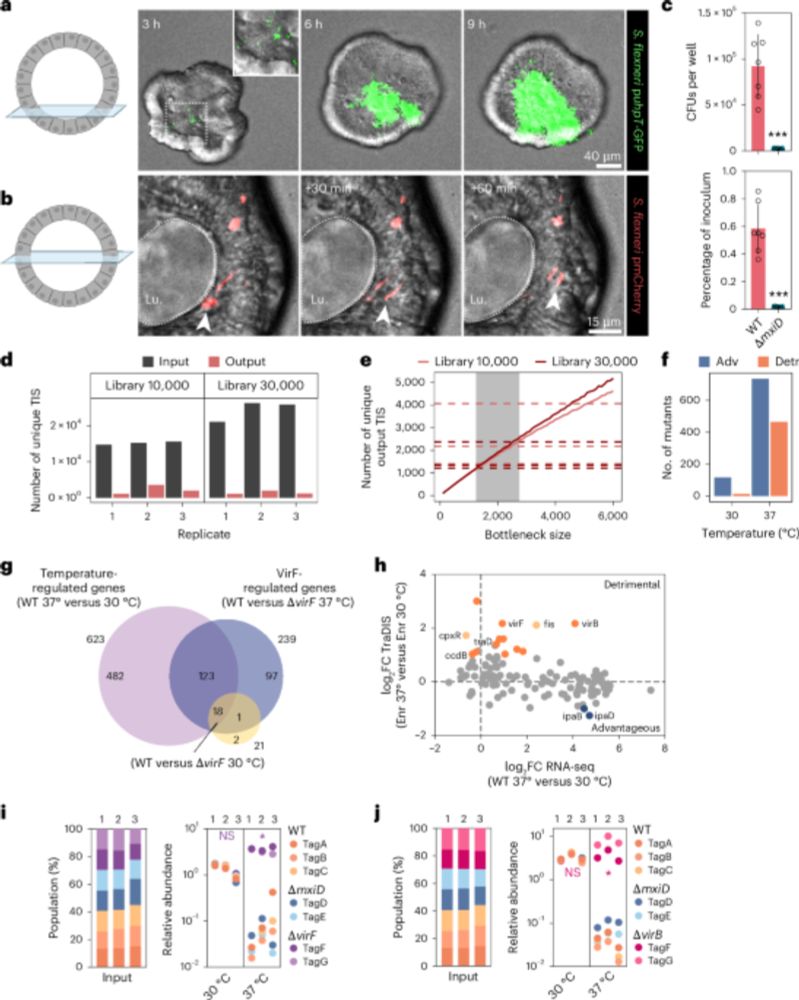
A scalable gut epithelial organoid model reveals the genome-wide colonization landscape of a human-adapted pathogen - Nature Genetics
A genome-wide screen using human gut epithelial organoids combined with transposon-directed insertion sequencing identifies over 100 Shigella flexneri genes required for epithelial colonization.
www.nature.com
Reposted by Maria Letizia Di Martino
Reposted by Maria Letizia Di Martino
SellinLab
@sellinlab.bsky.social
· May 30

Determinants of divergent Salmonella and Shigella epithelial colonization strategies resolved in human enteroids and colonoids | mBio
Pathogenic bacteria employ partially overlapping sets of virulence factor functions
to colonize host epithelia but can differ markedly in their pathogenesis and disease
kinetics. This work combines ba...
journals.asm.org