Lars Barquist
@lbarquist.bsky.social
4.5K followers
1.5K following
120 posts
Assistant prof @ UofT, associated scientist @ Helmholtz Institute for RNA-based Infection Research. Pathogen systems biology / informatics / functional genomics. Coastal New Hampshirite. Drink Moxie.
Posts
Media
Videos
Starter Packs
Pinned
Lars Barquist
@lbarquist.bsky.social
· Jun 12
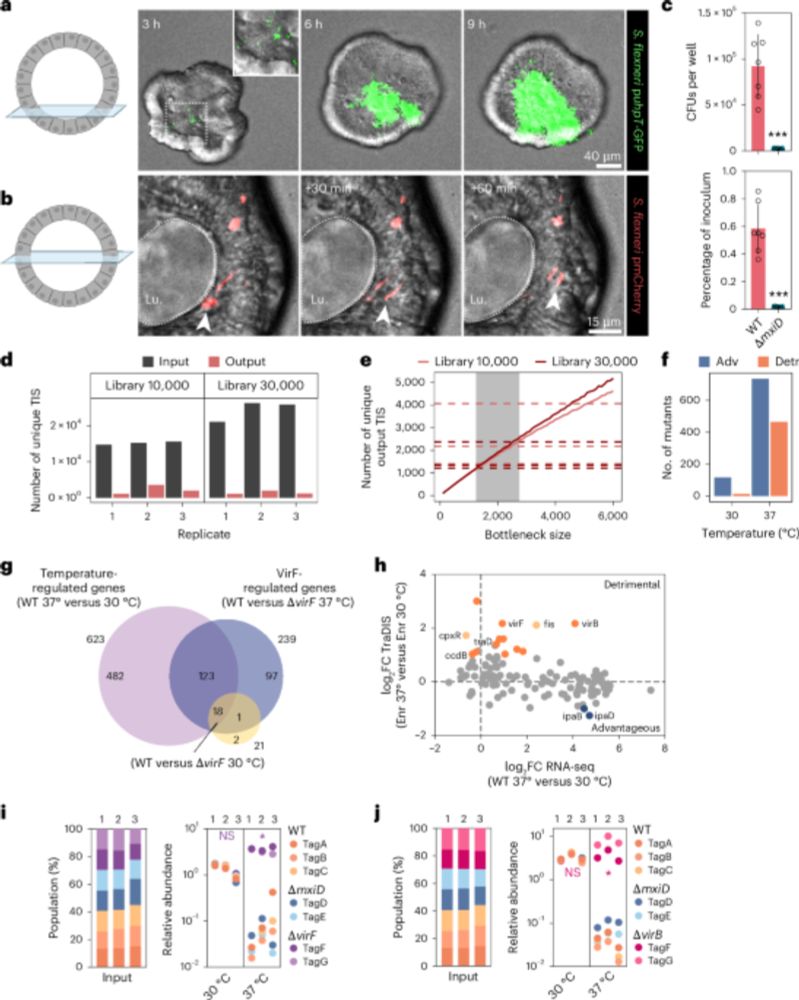
A scalable gut epithelial organoid model reveals the genome-wide colonization landscape of a human-adapted pathogen - Nature Genetics
A genome-wide screen using human gut epithelial organoids combined with transposon-directed insertion sequencing identifies over 100 Shigella flexneri genes required for epithelial colonization.
www.nature.com
Reposted by Lars Barquist
Reposted by Lars Barquist
Reposted by Lars Barquist
Reposted by Lars Barquist
Reposted by Lars Barquist
Reposted by Lars Barquist
Reposted by Lars Barquist
Reposted by Lars Barquist
James McInerney
@jomcinerney.bsky.social
· Aug 29

Genomic constraints shape the evolution of alternative routes to drug resistance in prokaryotes
Background Variation within the prokaryotic pangenome is not random, and natural selection that favours particular combinations of genes appears to dominate over random drift. What is less clear is wh...
www.biorxiv.org
Reposted by Lars Barquist