Arianne Babina
@ariannebabina.bsky.social
220 followers
290 following
15 posts
Lecturer in Bacteriology @ University of Glasgow | de novo evolution of bacterial gene regulatory networks | ✌️❤️RNA | she/her
Posts
Media
Videos
Starter Packs
Reposted by Arianne Babina
Reposted by Arianne Babina
Reposted by Arianne Babina
Reposted by Arianne Babina
Reposted by Arianne Babina
Reposted by Arianne Babina
Eduardo Rocha
@epcrocha.bsky.social
· Sep 8
Reposted by Arianne Babina
Reposted by Arianne Babina
Reposted by Arianne Babina
Reposted by Arianne Babina
Reposted by Arianne Babina
Reposted by Arianne Babina
Reposted by Arianne Babina
Reposted by Arianne Babina
Philip Ball
@philipcball.bsky.social
· Sep 5

Rethinking RNA-binding proteins: Riboregulation challenges prevailing views
The advent of system-wide proteomic approaches has largely expanded the number of
RNA-binding proteins (RBPs). This review discusses how recent discoveries are transforming
our understanding of biolog...
www.cell.com
Reposted by Arianne Babina
Philip Ball
@philipcball.bsky.social
· Sep 5

Rethinking RNA-binding proteins: Riboregulation challenges prevailing views
The advent of system-wide proteomic approaches has largely expanded the number of
RNA-binding proteins (RBPs). This review discusses how recent discoveries are transforming
our understanding of biolog...
www.cell.com
Reposted by Arianne Babina
James Connolly
@ruamicro.bsky.social
· Jul 30
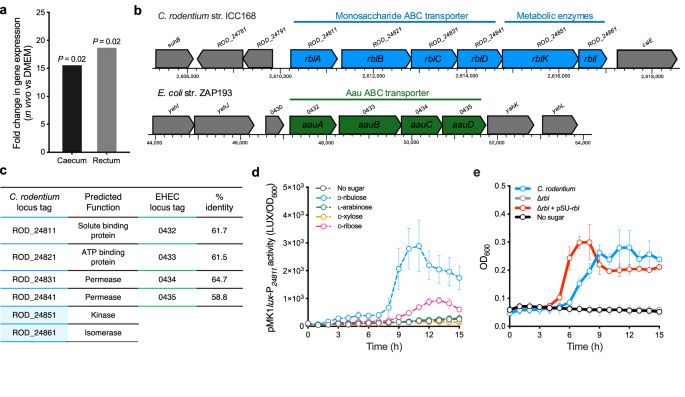
Convergent evolution of distinct D-ribulose utilisation pathways in attaching and effacing pathogens - Nature Communications
Cottam et al. identify distinct pathways for D-ribulose utilisation in pathogenic Escherichia coli and Citrobacter rodentium, providing mechanistic details and suggesting convergent evolution towards ...
www.nature.com
Reposted by Arianne Babina
Will Smith
@willpjsmith.bsky.social
· Aug 31

Type VI secretion system activity at lethal antibiotic concentrations leads to overestimation of weapon potency
Competition assays are a mainstay of modern microbiology, offering a simple and cost-effective means to quantify microbe–microbe interactions in vitro. Here, we demonstrate a key weakness of this meth...
www.microbiologyresearch.org












