Mikael Marttinen
@mmarttinen.bsky.social
59 followers
71 following
11 posts
Postdoc researching gene regulation in disease || Tampere University and EMBL
Posts
Media
Videos
Starter Packs
Pinned
Mikael Marttinen
@mmarttinen.bsky.social
· May 26
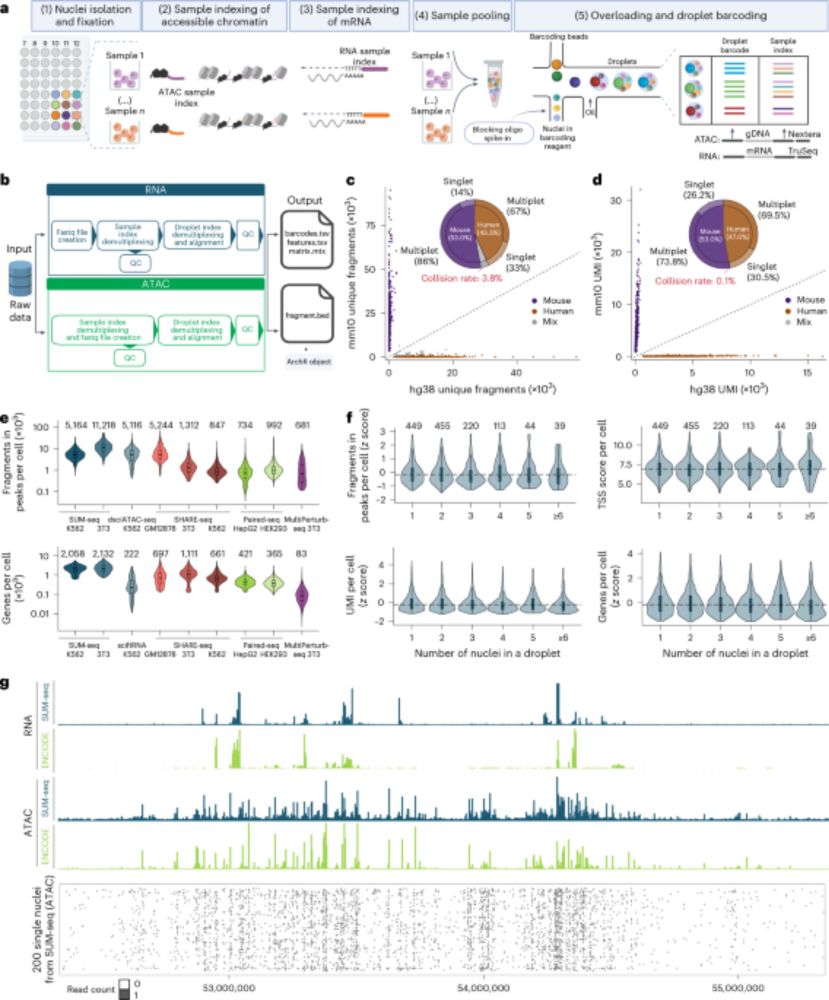
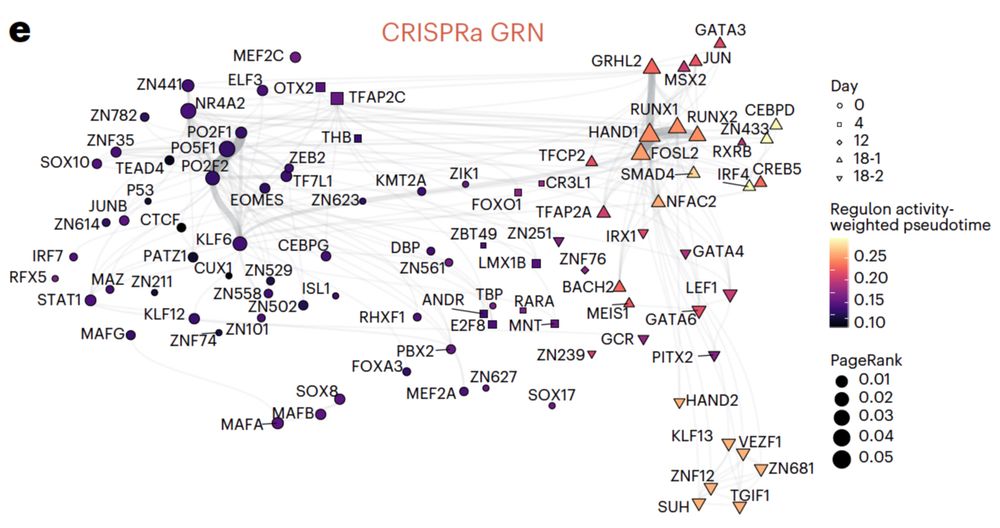
Single-cell ultra-high-throughput multiplexed chromatin and RNA profiling reveals gene regulatory dynamics - Nature Methods
This work presents SUM-seq, an ultra-high-throughput method for co-profiling chromatin accessibility and gene expression in single nuclei across multiplexed samples, advancing the study of gene regula...
doi.org
Reposted by Mikael Marttinen
Mikael Marttinen
@mmarttinen.bsky.social
· May 26
Mikael Marttinen
@mmarttinen.bsky.social
· May 26
Mikael Marttinen
@mmarttinen.bsky.social
· May 26

Single-cell ultra-high-throughput multiplexed chromatin and RNA profiling reveals gene regulatory dynamics - Nature Methods
This work presents SUM-seq, an ultra-high-throughput method for co-profiling chromatin accessibility and gene expression in single nuclei across multiplexed samples, advancing the study of gene regula...
doi.org
Reposted by Mikael Marttinen
Reposted by Mikael Marttinen