Nacho Molina
@molinalab.bsky.social
1.6K followers
1.2K following
100 posts
Group leader of the Stochastic Systems Biology Lab at IGBMC - CNRS - University of Strasbourg. Models of gene regulation based on biophysics-informed deep learning: https://www.igbmc.fr/molina
Posts
Media
Videos
Starter Packs
Pinned
Nacho Molina
@molinalab.bsky.social
· Aug 21
Reposted by Nacho Molina
Reposted by Nacho Molina
Reposted by Nacho Molina
Reposted by Nacho Molina
Yad Ghavi-Helm
@yghavi.bsky.social
· Jul 23
Reposted by Nacho Molina
Reposted by Nacho Molina
Reposted by Nacho Molina
Reposted by Nacho Molina
Nacho Molina
@molinalab.bsky.social
· May 29
Reposted by Nacho Molina
Nacho Molina
@molinalab.bsky.social
· May 22
Reposted by Nacho Molina
Lars Velten
@larsplus.bsky.social
· May 21
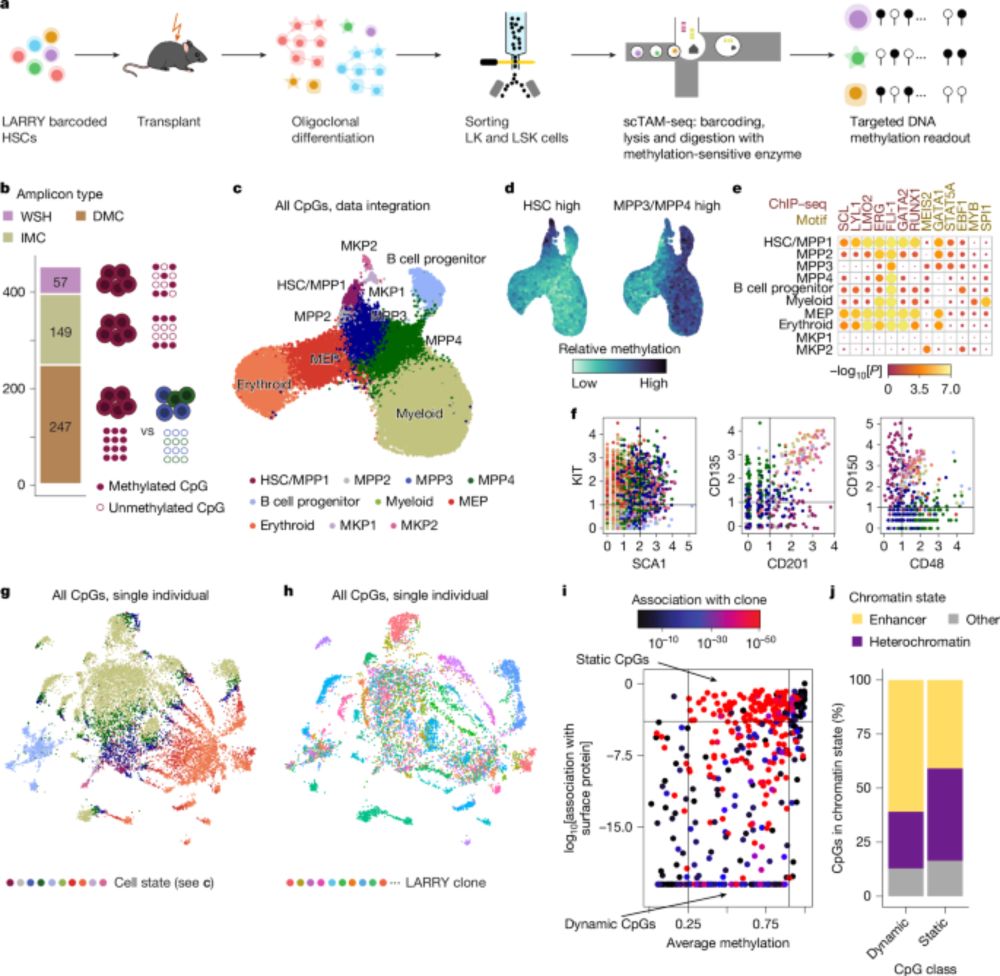
Clonal tracing with somatic epimutations reveals dynamics of blood ageing - Nature
The discovery that DNA methylation of different CpG sites can serve as digital barcodes of clonal identity led to the development of EPI-Clone, an algorithm that enables single-cell lineage tracing th...
doi.org
Reposted by Nacho Molina
Nacho Molina
@molinalab.bsky.social
· May 20
Reposted by Nacho Molina
Selin Jessa
@selinjessa.bsky.social
· May 3

Dissecting regulatory syntax in human development with scalable multiomics and deep learning
Transcription factors (TFs) establish cell identity during development by binding regulatory DNA in a sequence-specific manner, often promoting local chromatin accessibility, and regulating gene expre...
www.biorxiv.org
Nacho Molina
@molinalab.bsky.social
· May 19
Reposted by Nacho Molina
Nacho Molina
@molinalab.bsky.social
· May 19