Selin Jessa
@selinjessa.bsky.social
920 followers
1.3K following
39 posts
(she/her) Computational biologist and post-doc scientist in the Greenleaf and Kundaje labs at Stanford. Interested in understanding how cells know what to become (transcription factors, gene regulation, dev bio, open science) www.selinjessa.com
Posts
Media
Videos
Starter Packs
Pinned
Selin Jessa
@selinjessa.bsky.social
· May 3

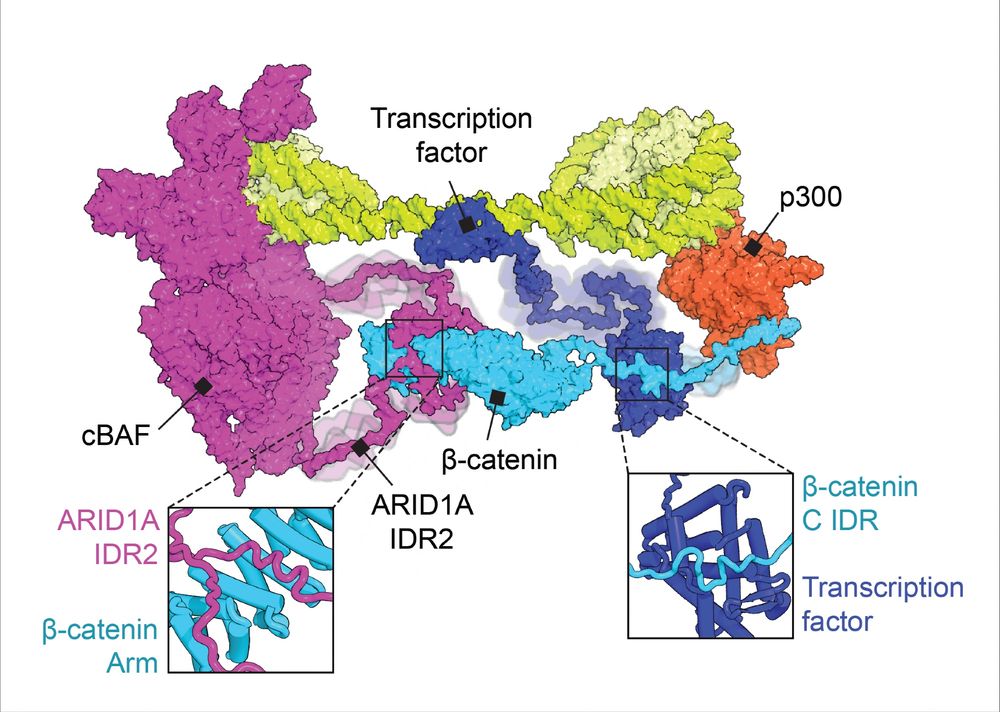
Dissecting regulatory syntax in human development with scalable multiomics and deep learning
Transcription factors (TFs) establish cell identity during development by binding regulatory DNA in a sequence-specific manner, often promoting local chromatin accessibility, and regulating gene expre...
www.biorxiv.org
Reposted by Selin Jessa
Reposted by Selin Jessa
Reposted by Selin Jessa
Reposted by Selin Jessa
Reposted by Selin Jessa
Reposted by Selin Jessa
Reposted by Selin Jessa
Reposted by Selin Jessa
Benoit Bruneau
@benoitbruneau.bsky.social
· Aug 20

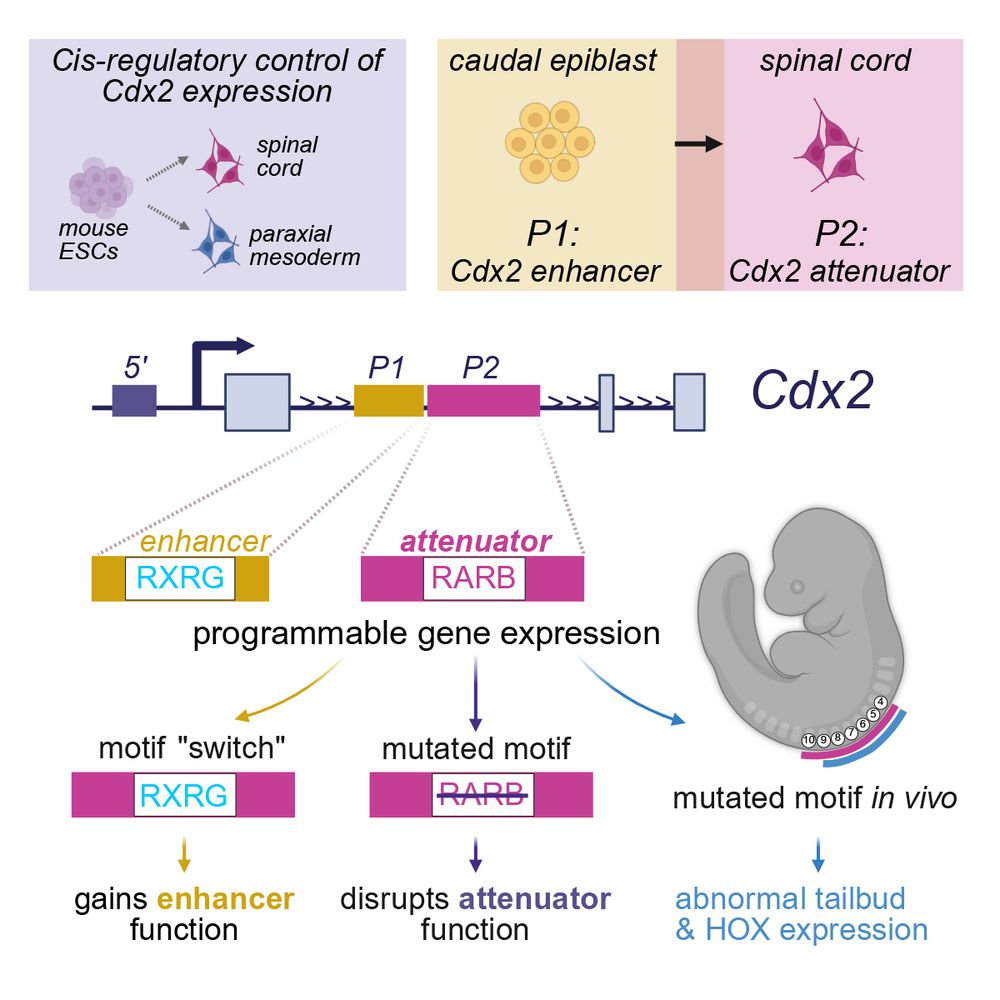
Reduced TBX5 dosage undermines developmental control of atrial cardiomyocyte identity in a model of human atrial disease
There is evidence for a shared genetic basis of atrial septal defects (ASDs) and atrial fibrillation (AF), but it remains unclear how genetic susceptibility leads to these distinct human atrial diseas...
www.biorxiv.org
Reposted by Selin Jessa
María Mariner
@mariamafau.bsky.social
· Jul 24
Reposted by Selin Jessa
Reposted by Selin Jessa
Reposted by Selin Jessa
Reposted by Selin Jessa
Doug Fowler
@dougfowler.bsky.social
· Jul 21
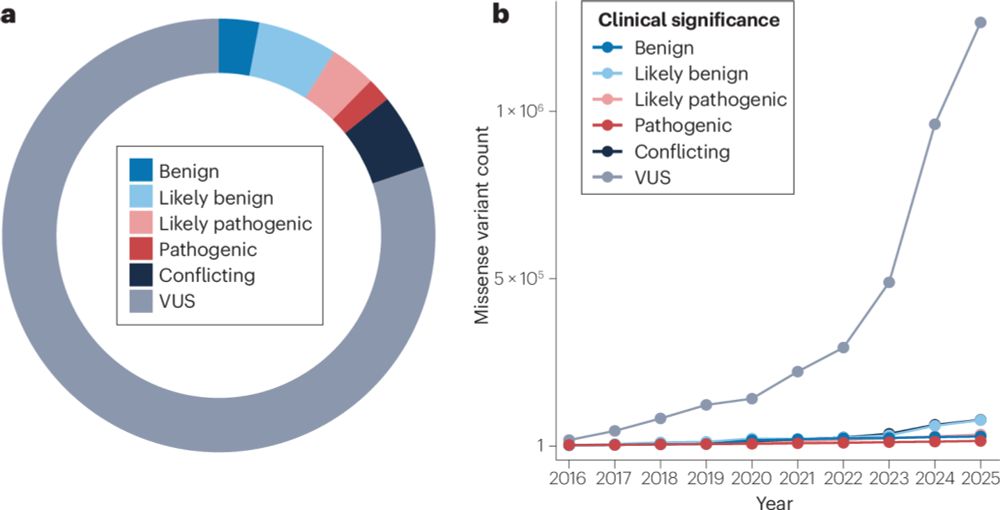
Multiplexed assays of variant effect for clinical variant interpretation
Nature Reviews Genetics - Multiplexed assays of variant effect (MAVEs) are highly scalable experimental approaches used to generate functional data for genetic variants. In this Review, McEwen et...
www.nature.com
Reposted by Selin Jessa
Li Zhao
@lizhao.bsky.social
· Jul 15
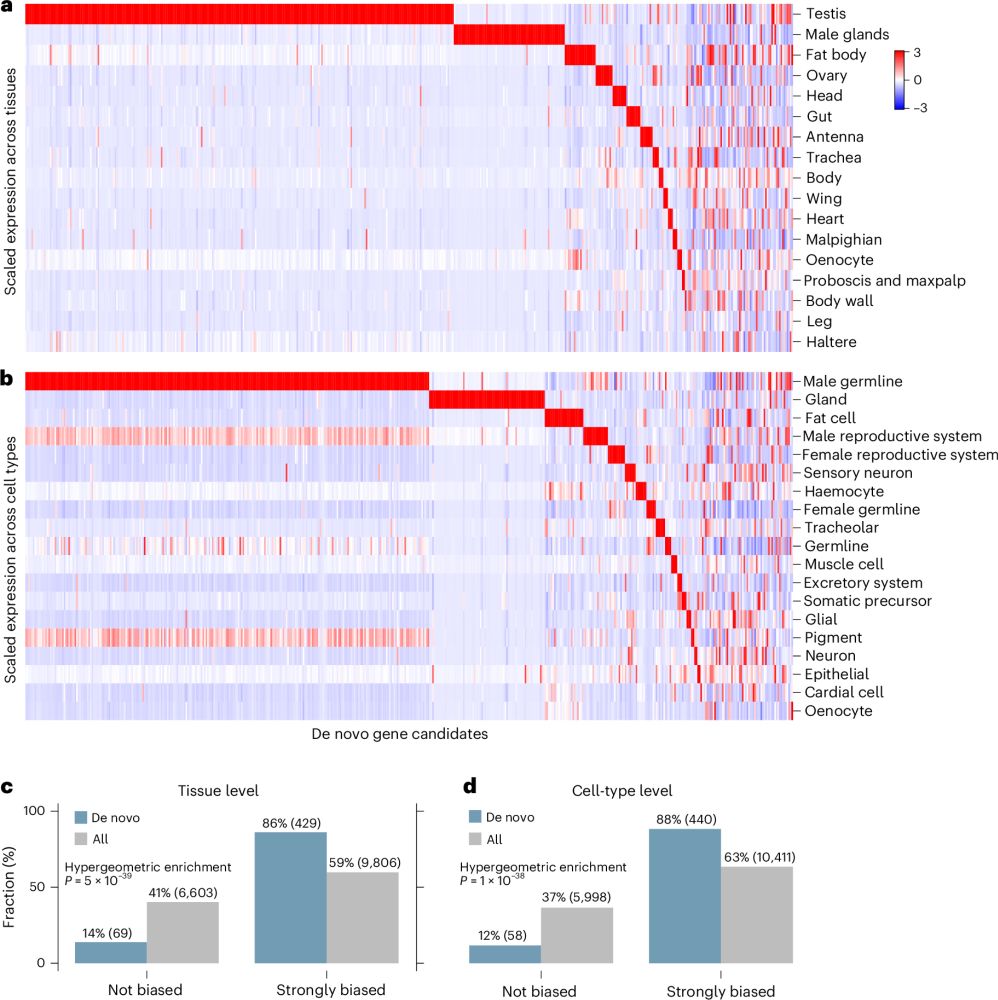
Gene regulatory networks and essential transcription factors for de novo-originated genes
Nature Ecology & Evolution - A combination of computational methods applied to single-cell RNA sequencing data and genetic experiments shows that a small number of transcription factors are...
rdcu.be
Reposted by Selin Jessa
Reposted by Selin Jessa
Reposted by Selin Jessa
Žiga Avsec
@avsecz.bsky.social
· Jun 25
Reposted by Selin Jessa
Žiga Avsec
@avsecz.bsky.social
· Jun 25
Reposted by Selin Jessa
Reposted by Selin Jessa
Reposted by Selin Jessa
Pooja Kathail
@poojakathail.bsky.social
· Nov 20

Leveraging genomic deep learning models for non-coding variant effect prediction
The majority of genetic variants identified in genome-wide association studies of complex traits are non-coding, and characterizing their function remains an important challenge in human genetics. Gen...
arxiv.org