The Lewis Lab
@peterlewislab.bsky.social
930 followers
710 following
69 posts
Chromatin biochemistry and genomics in development and cancer. UW-Madison School of Medicine and Public Health
TheLewisLab.net
Posts
Media
Videos
Starter Packs
Pinned
Reposted by The Lewis Lab
Reposted by The Lewis Lab
Reposted by The Lewis Lab
Reposted by The Lewis Lab
Reposted by The Lewis Lab
Reposted by The Lewis Lab
Reposted by The Lewis Lab
Reposted by The Lewis Lab
AmélieFT@AFTLab
@amelieft.bsky.social
· Aug 13
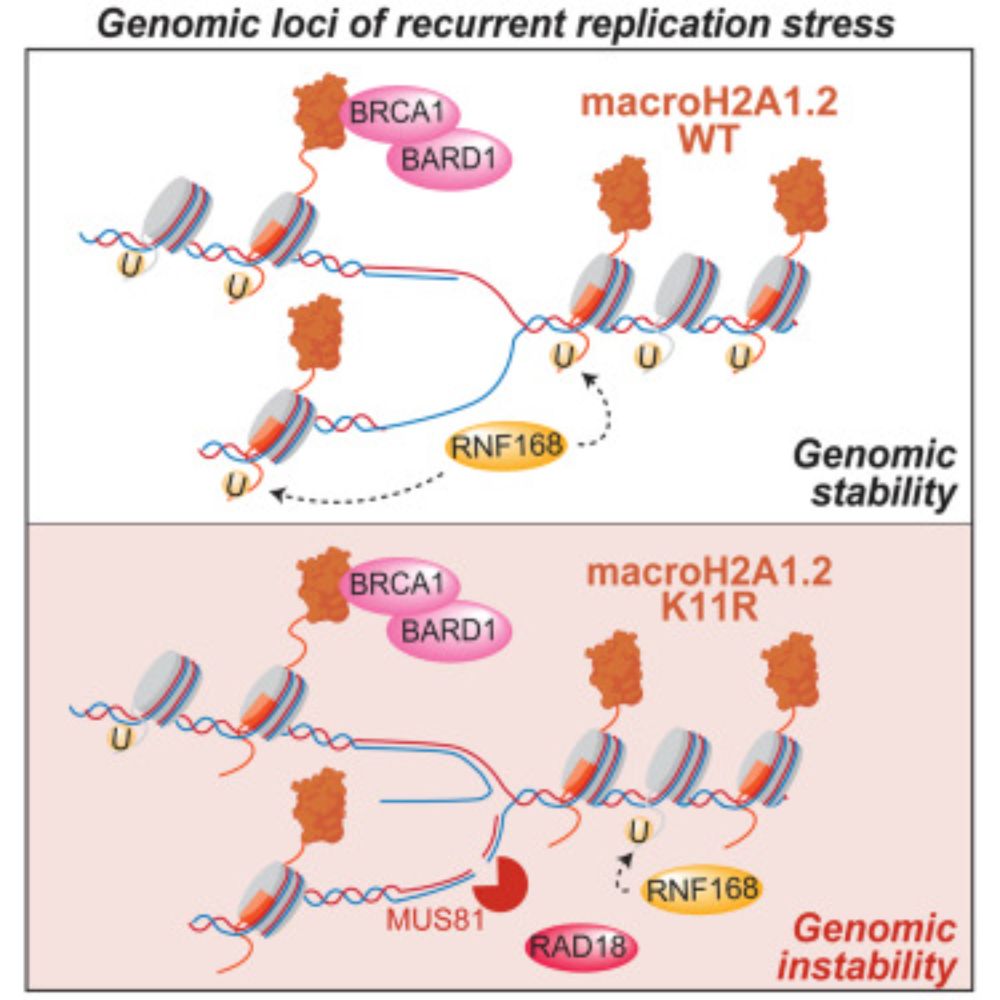
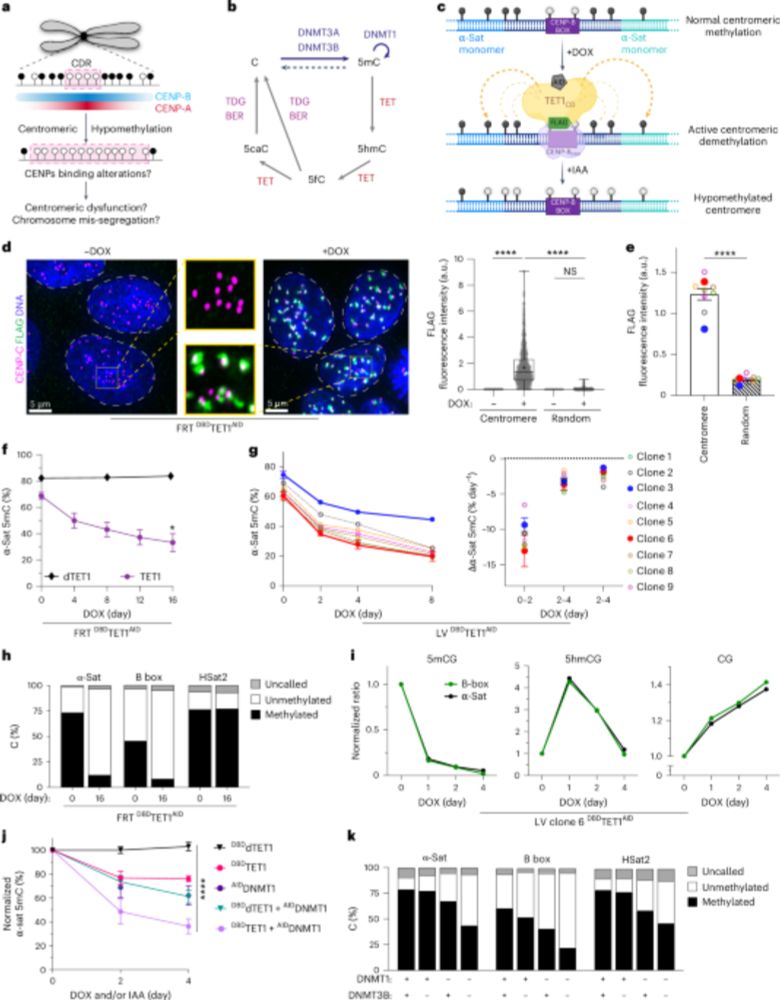
Ubiquitination of the histone variant mH2A1.2 prevents toxic RAD18 accumulation at a subset of genomic loci upon replication stress
Using biochemical assays and a mutant disrupting RNF168-dependent ubiquitination of
the histone variant macroH2A1.2, Galloy et al. identified an unexpected role for histone
mH2A1.2 ubiquitination in p...
www.cell.com
Reposted by The Lewis Lab
Abdel Mazouzi
@amazouzi.bsky.social
· Aug 13
Reposted by The Lewis Lab
Katherine Aird
@airdlab.bsky.social
· Aug 14

The chemotherapy-induced senescence-associated secretome promotes cell detachment and metastatic dissemination through metabolic reprogramming
Cellular senescence, characterized by a stable cell cycle arrest, is a well-documented consequence of several widely used chemotherapeutics that has context-dependent roles in cancer. Although senesce...
www.biorxiv.org
Reposted by The Lewis Lab
Reposted by The Lewis Lab
Schubeler Lab
@schubelerlab.bsky.social
· Aug 12

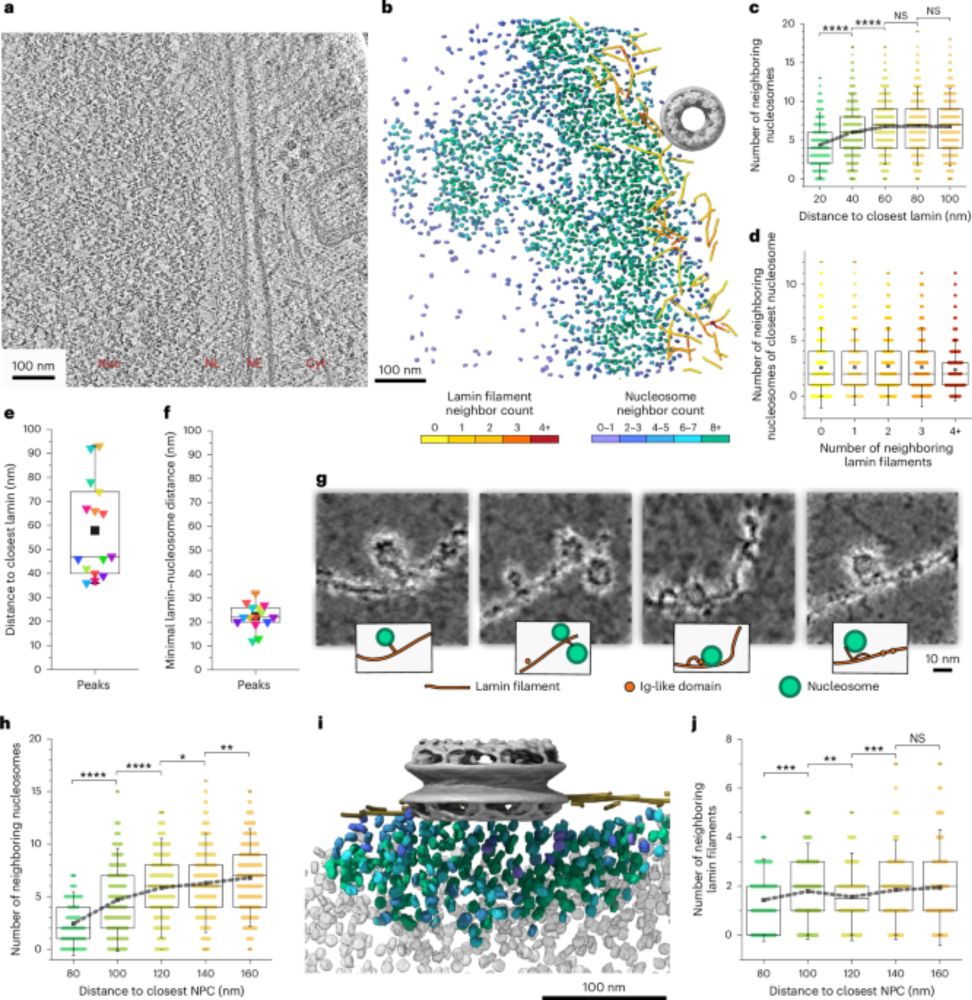
A novel deep learning-based framework reveals a continuum of chromatin sensitivities across transcription factors
The genome-wide binding of many transcription factors (TFs) depends not only on the presence of their recognition motifs, but also on the surrounding chromatin context. This raises the question of how...
www.biorxiv.org
The Lewis Lab
@peterlewislab.bsky.social
· Aug 13
The Lewis Lab
@peterlewislab.bsky.social
· Aug 13
Reposted by The Lewis Lab