Johannes Morstein
@morstein.bsky.social
1.4K followers
790 following
15 posts
Assistant Professor at Caltech | Chemical Biologist
morsteinlab.caltech.edu
Posts
Media
Videos
Starter Packs
Reposted by Johannes Morstein
Johannes Morstein
@morstein.bsky.social
· Aug 21
Reposted by Johannes Morstein
Reposted by Johannes Morstein
Itay Budin
@ibudin.bsky.social
· Aug 15

Organelle-Targeted Laurdans Measure Heterogeneity in Subcellular Membranes and Their Responses to Saturated Lipid Stress
Organelles feature characteristic lipid compositions that lead to differences in membrane properties. In cells, membrane ordering and fluidity are commonly measured using the solvatochromic dye Laurdan, whose fluorescence is sensitive to lipid packing. As a general lipophilic dye, Laurdan stains all hydrophobic environments in cells; therefore, it is challenging to characterize membrane properties in specific organelles or assess their responses to pharmacological treatments in intact cells. Here, we describe the synthesis and application of Laurdan-derived probes that read out the membrane packing of individual cellular organelles. The set of organelle-targeted Laurdans (OTL) localizes to the ER, mitochondria, lysosomes, and Golgi compartments with high specificity while retaining the spectral resolution needed to detect biological changes in membrane ordering. We show that ratiometric imaging with OTLs can resolve membrane heterogeneity within organelles as well as changes in lipid packing resulting from inhibition of trafficking or bioenergetic processes. We apply these probes to characterize organelle-specific responses to saturated lipid stress. While the ER and lysosomal membrane fluidity is sensitive to exogenous saturated fatty acids, that of mitochondrial membranes is protected. We then use differences in ER membrane fluidity to sort populations of cells based on their fatty acid diet, highlighting the ability of organelle-localized solvatochromic probes to distinguish between cells based on their metabolic state. These results expand the repertoire of targeted membrane probes and demonstrate their application in interrogating lipid dysregulation.
pubs.acs.org
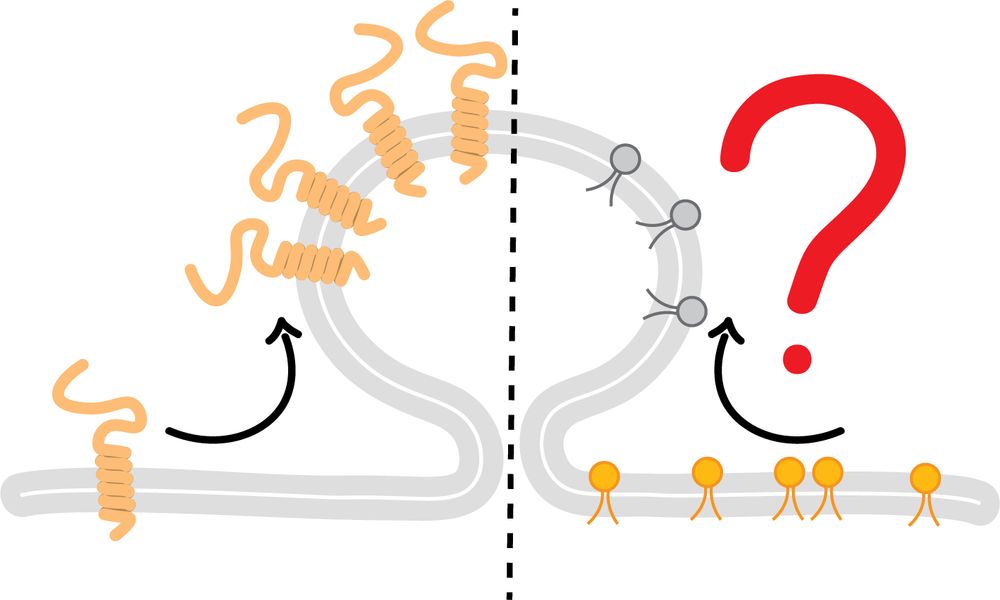
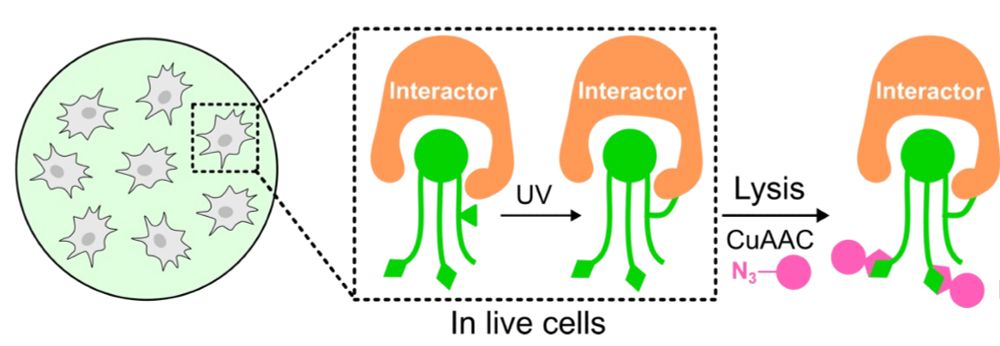
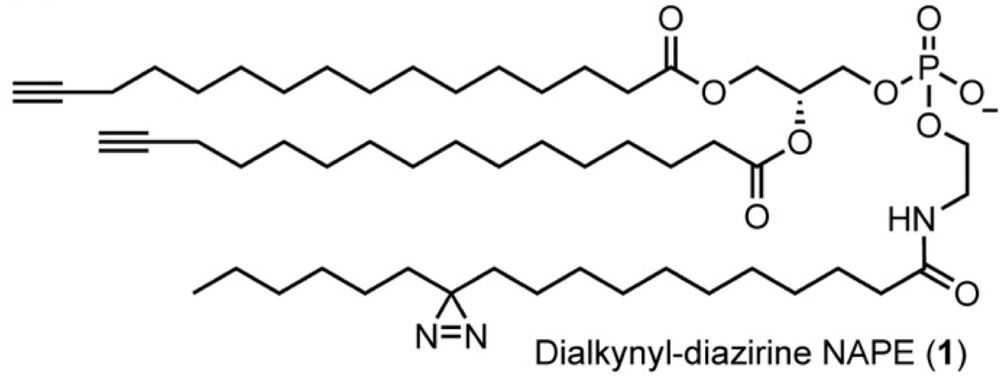
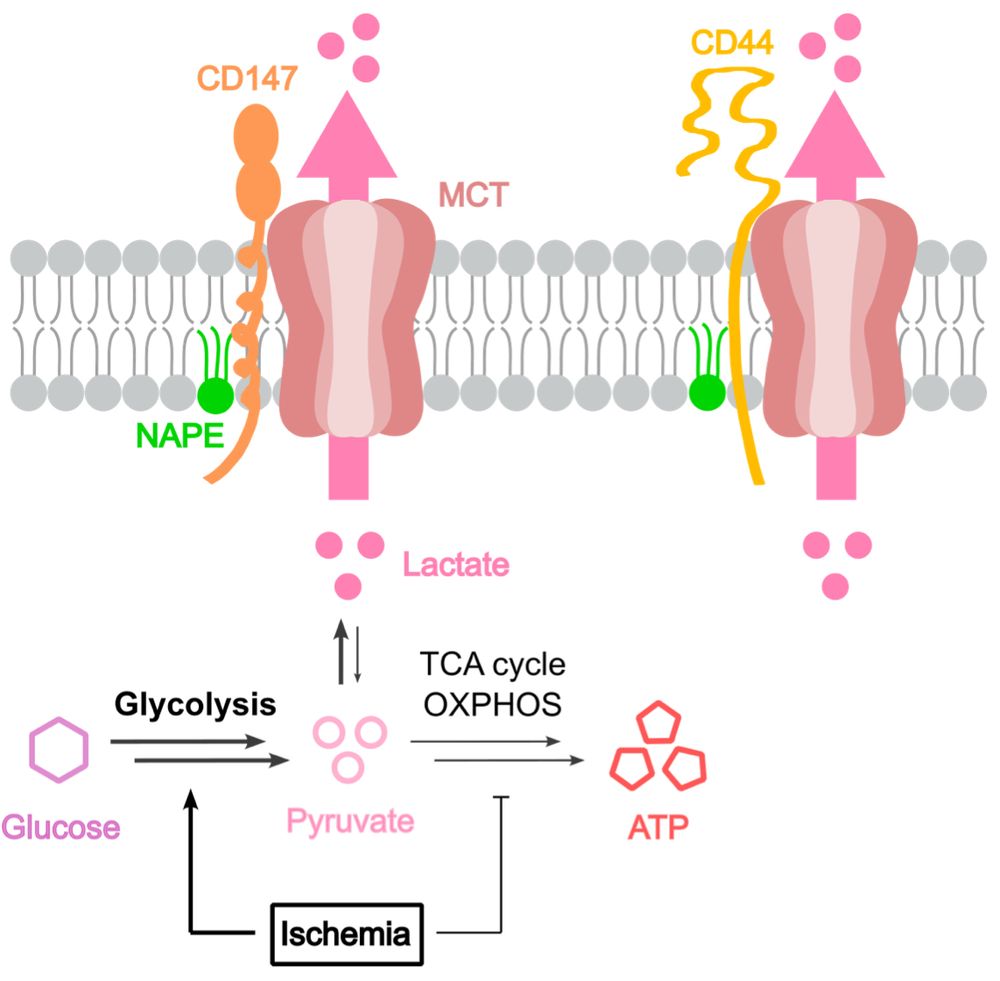
Johannes Morstein
@morstein.bsky.social
· Aug 14
Johannes Morstein
@morstein.bsky.social
· Aug 14

Optical Control of Membrane Viscosity Modulates ER-to-Golgi Trafficking
The lipid composition of cellular membranes is highly dynamic and undergoes continuous remodeling, affecting the biophysical properties critical to biological function. Here, we introduce an optical a...
pubs.acs.org
Reposted by Johannes Morstein
Judi Simcox
@judisimcox.bsky.social
· May 15
Reposted by Johannes Morstein
Johannes Morstein
@morstein.bsky.social
· Mar 27
Johannes Morstein
@morstein.bsky.social
· Mar 26
Reposted by Johannes Morstein
Reposted by Johannes Morstein
Reposted by Johannes Morstein
Dirk Trauner
@dirktrauner.bsky.social
· Jan 15

Concise Synthesis of (−)-Veratramine and (−)-20-iso-Veratramine via Aromative Diels–Alder Reaction
A concise and convergent synthesis of the isosteroidal alkaloids veratramine and 20-iso-veratramine has been accomplished. A Horner–Wadsworth–Emmons olefination joins two chiral building blocks of app...
pubs.acs.org
Reposted by Johannes Morstein
Reposted by Johannes Morstein
Reposted by Johannes Morstein
Chris Chang
@christhechang.bsky.social
· Nov 27
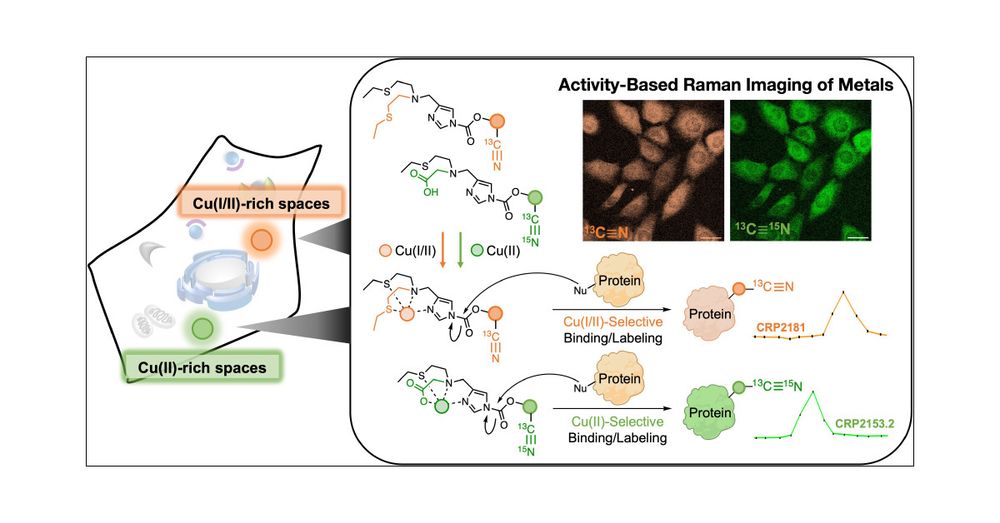
An Activity-Based Sensing Approach to Multiplex Mapping of Labile Copper Pools by Stimulated Raman Scattering
Molecular imaging with analyte-responsive probes offers a powerful chemical approach to studying biological processes. Many reagents for bioimaging employ a fluorescence readout, but the relatively br...
pubs.acs.org
Johannes Morstein
@morstein.bsky.social
· Nov 26