Matthew Williams
@mp-williams.bsky.social
200 followers
97 following
6 posts
Archaeogeneticist by day and night. Postdoc in the Huber Lab at Penn State. Lover of the ancient Near East, philosophy, and coffee.
Posts
Media
Videos
Starter Packs
Reposted by Matthew Williams
Reposted by Matthew Williams
Tom Booth
@boothicus.bsky.social
· Aug 26

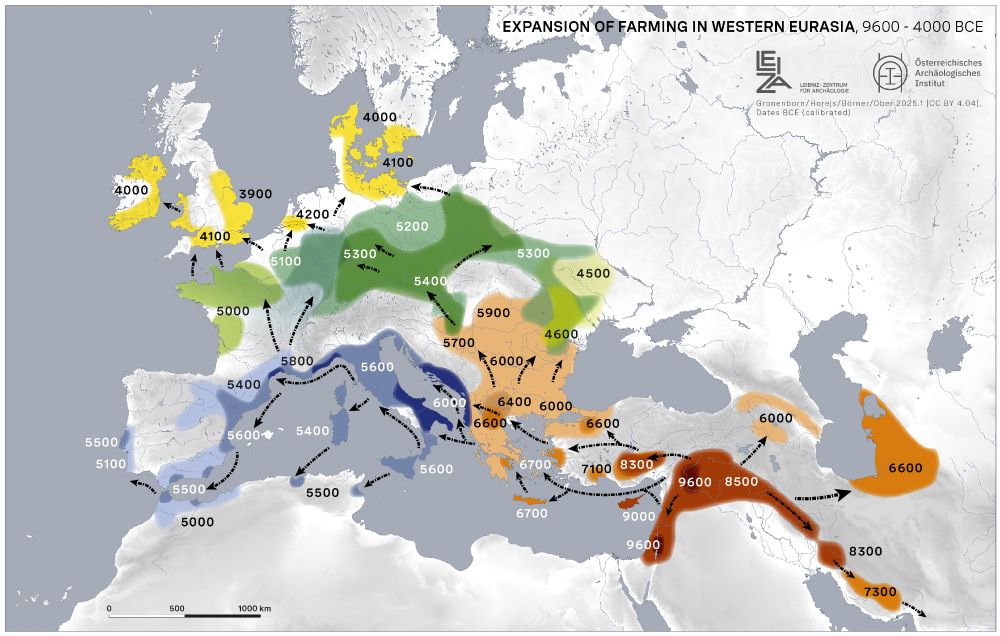
Modeling the European Neolithic expansion suggests predominant within-group mating and limited cultural transmission - Nature Communications
The Neolithic Revolution marked an important shift from foraging to farming in human society. Here, the authors show that in Europe the spread of farming involved mostly within-group mating and limite...
www.nature.com
Matthew Williams
@mp-williams.bsky.social
· Aug 27
Reposted by Matthew Williams
Rosi Fregel
@rfregel.bsky.social
· Aug 21

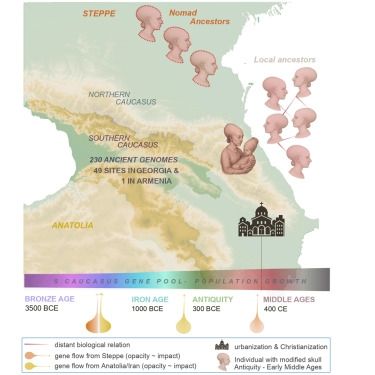
The AADR Visualizer: An ArcGIS Online Visualizer for ancient human DNA from the Allen Ancient DNA Resource
AbstractMotivation. The AADR Visualizer is designed to be a public, user-friendly, web-based graphical user interface for visualizing and filtering ancient
academic.oup.com
Reposted by Matthew Williams
Reposted by Matthew Williams
Reposted by Matthew Williams
Stacey Ogden 🦔
@labogden.bsky.social
· Aug 11
NOT-OD-25-138: Request for Information on Maximizing Research Funds by Limiting Allowable Publishing Costs
NIH Funding Opportunities and Notices in the NIH Guide for Grants and Contracts: Request for Information on Maximizing Research Funds by Limiting Allowable Publishing Costs NOT-OD-25-138. NIH
grants.nih.gov
Reposted by Matthew Williams
Stephen Hill
@srhastraea.bsky.social
· Aug 9
Claus Wilke
@clauswilke.com
· Aug 9

PhD-level intelligence or the graduate student from hell
AI companies love to tout that their models are approaching—or have reached—PhD-level intelligence. This is blatant nonsensical marketing geared towards an audience that deeply misunderstands what a P...
clauswilke.substack.com
Reposted by Matthew Williams
Reposted by Matthew Williams
Reposted by Matthew Williams
Reposted by Matthew Williams
Reposted by Matthew Williams
Reposted by Matthew Williams
Skyler Berardi
@skylerberardi.bsky.social
· Jul 31
Reposted by Matthew Williams
Reposted by Matthew Williams
Marco Capodiferro
@marcocapo.bsky.social
· Jul 24

Archaic ancestry inference in imputed ancient human genomes
When modern humans expanded from Africa into Eurasia, they interbred with archaic hominins such as Neanderthals and Denisovans. This groundbreaking discovery, made in part possible through the genomic...
doi.org
Reposted by Matthew Williams
Reposted by Matthew Williams
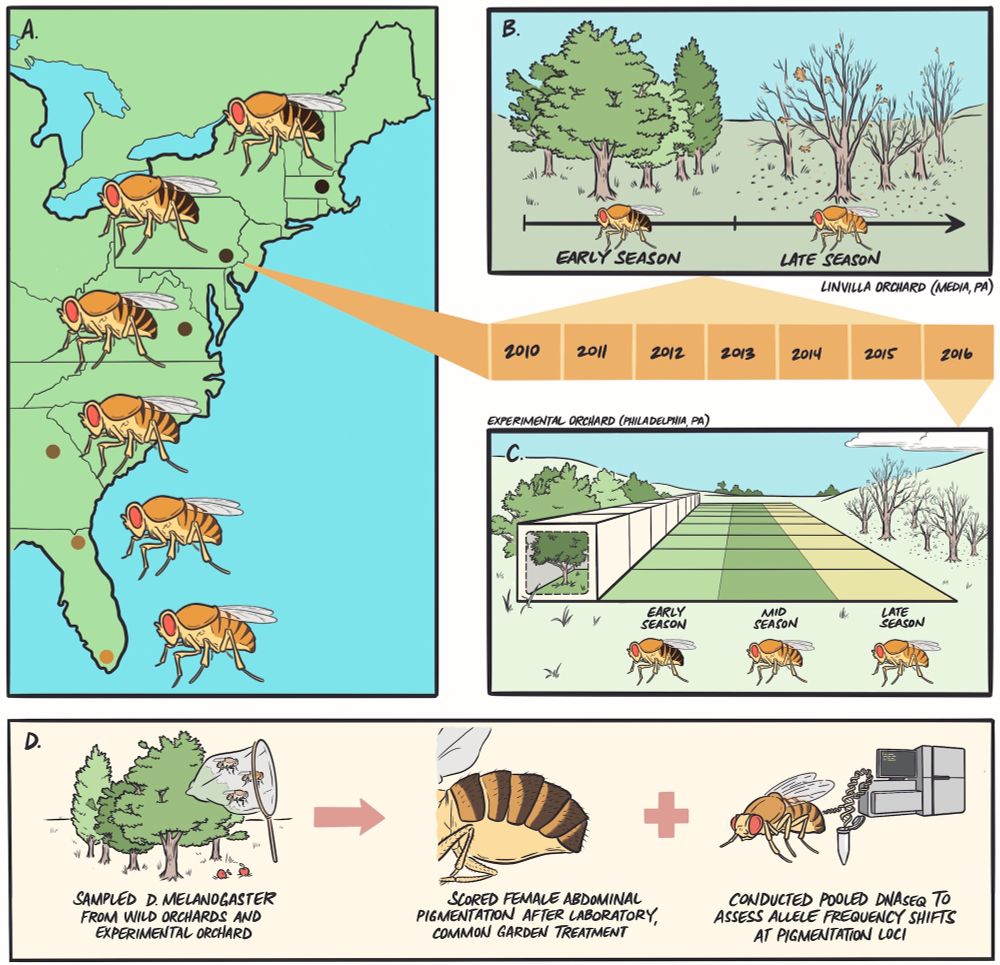
EvolDir
@evoldir.bsky.social
· Jul 24

We are a computational lab focusing on developing machine learning algorithms for biological data. Currently we are working on inference tasks related to natural selection and demographic inference, as well as generative models for genomic data from humans, mosquitos, and other species.
saramathieson.github.io
Reposted by Matthew Williams
Reposted by Matthew Williams
Hanbin Lee
@epigenci.bsky.social
· Jul 20
Reposted by Matthew Williams
Hanbin Lee
@epigenci.bsky.social
· Jul 20
Reposted by Matthew Williams
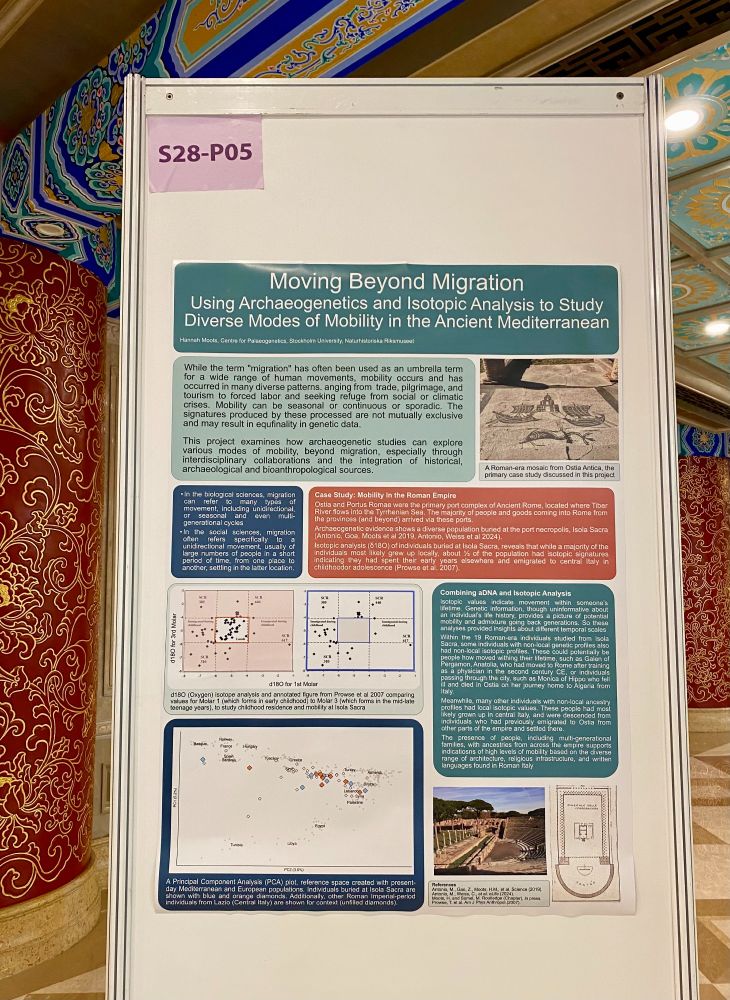
Hannah Moots
@mootspoints.bsky.social
· Jul 20