Mitchell R. Vollger
@mrvollger.bsky.social
240 followers
150 following
13 posts
https://mrvollger.github.io
Posts
Media
Videos
Starter Packs
Pinned
Reposted by Mitchell R. Vollger
Reposted by Mitchell R. Vollger
Reposted by Mitchell R. Vollger
Reposted by Mitchell R. Vollger
Daniela C. Soto
@dcsoto.bsky.social
· Jul 21
Reposted by Mitchell R. Vollger
Reposted by Mitchell R. Vollger
Reposted by Mitchell R. Vollger
Reposted by Mitchell R. Vollger
Jenna Norton
@jenna-m-norton.bsky.social
· Jun 10
Reposted by Mitchell R. Vollger
Reposted by Mitchell R. Vollger
Alex Hoischen
@ahoischen.bsky.social
· May 26
Reposted by Mitchell R. Vollger
Rajiv McCoy
@rajivmccoy.bsky.social
· Mar 3

A T2T-CHM13 recombination map and globally diverse haplotype reference panel improves phasing and imputation
The T2T-CHM13 complete human reference genome contains ~200 Mb of newly resolved sequence, improving read mapping and variant calling compared to GRCh38. However, the benefits of using complete refere...
www.biorxiv.org
Reposted by Mitchell R. Vollger
Megan Dennis
@mydennis.bsky.social
· Feb 5
Reposted by Mitchell R. Vollger
Adam Phillippy
@aphillippy.bsky.social
· Jan 29
Eric Topol
@erictopol.bsky.social
· Jan 29
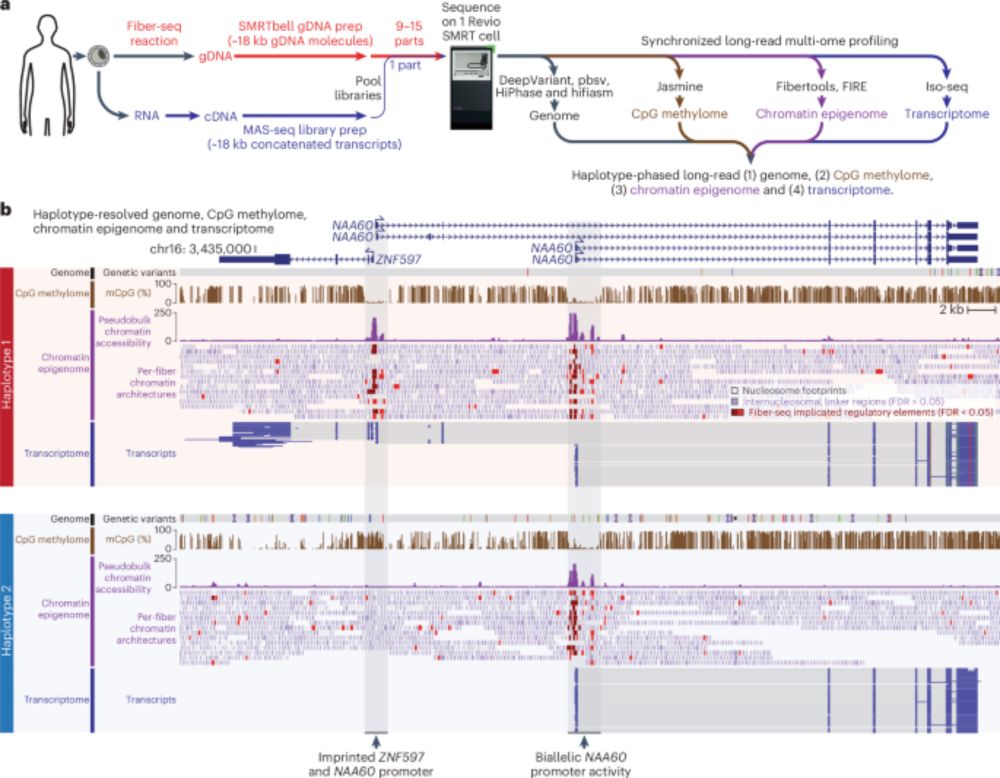
Synchronized long-read genome, methylome, epigenome and transcriptome profiling resolve a Mendelian condition - Nature Genetics
Simultaneous profiling of the genome, methylome, epigenome and transcriptome using single-molecule chromatin fiber sequencing and multiplexed arrays isoform sequencing identifies the genetic and molec...
www.nature.com
Reposted by Mitchell R. Vollger
Eric Topol
@erictopol.bsky.social
· Jan 29

Synchronized long-read genome, methylome, epigenome and transcriptome profiling resolve a Mendelian condition - Nature Genetics
Simultaneous profiling of the genome, methylome, epigenome and transcriptome using single-molecule chromatin fiber sequencing and multiplexed arrays isoform sequencing identifies the genetic and molec...
www.nature.com
Reposted by Mitchell R. Vollger
Megan Dennis
@mydennis.bsky.social
· Jan 9

HHMI Names the 2024 Hanna Gray Fellows | HHMI
The cohort includes early career scientists working in research areas ranging from treatment-resistant cancers to how animals evolved to live on land. The HHMI Hanna H. Gray Fellows Program provides e...
www.hhmi.org





