Posts
Media
Videos
Starter Packs
Reposted by Mile Sikic
Yuichi Shiraishi
@friend1ws.bsky.social
· Jul 30

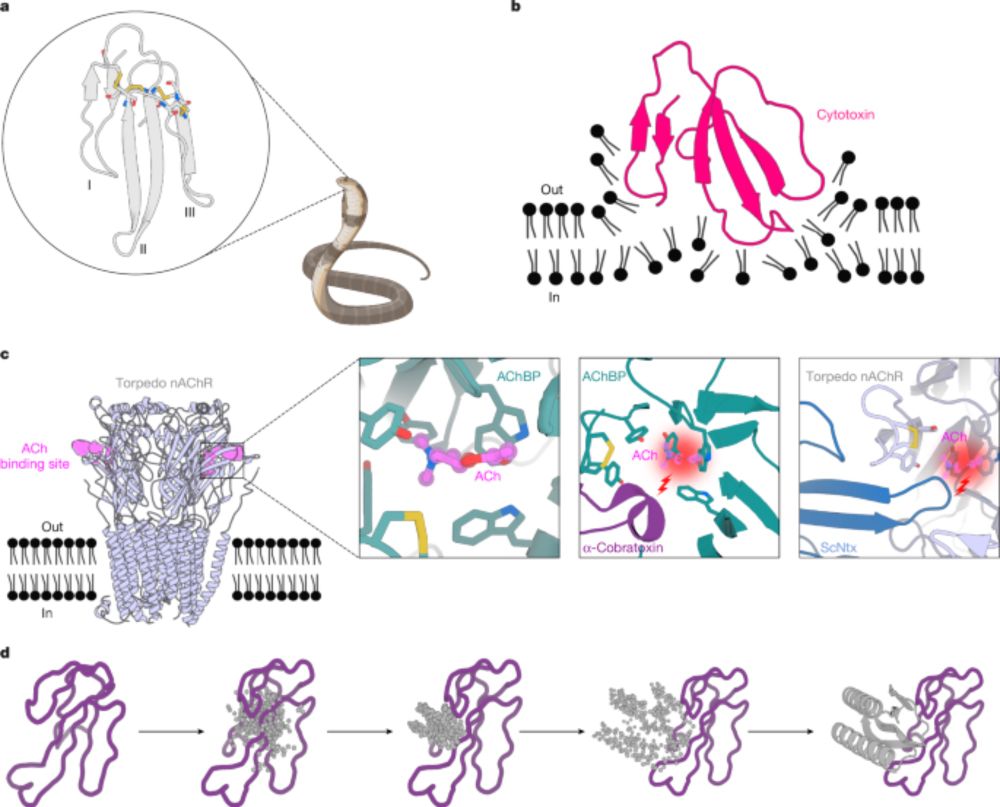
Rare k-mers reveal centromere haplogroups underlying human diversity and cancer translocations
Centromeres are among the most diverse and dynamically evolving regions of the human genome and are commonly affected in various human cancers. However, organized into highly repetitive α-satellite hi...
biorxiv.org
Mile Sikic
@msikic.bsky.social
· Aug 9
Mile Sikic
@msikic.bsky.social
· Jul 19

A Complete Telomere-to-Telomere Diploid Reference Genome for Indian Population
Human reference genomes have been instrumental in advancing genomic and biomedical research, but South and Southeast Asian populations are underrepresented, despite accounting for a large proportion o...
biorxiv.org
Mile Sikic
@msikic.bsky.social
· Jul 7

RiNALMo: general-purpose RNA language models can generalize well on structure prediction tasks - Nature Communications
RiNALMo, a large-scale RNA language model trained on non-coding RNA sequences, captures structural information and achieves state-of-the-art performance on multiple tasks, notably generalizing to unse...
nature.com
Mile Sikic
@msikic.bsky.social
· May 17
Reposted by Mile Sikic
Reposted by Mile Sikic
Mile Sikic
@msikic.bsky.social
· Apr 14
Reposted by Mile Sikic
Reposted by Mile Sikic
Mile Sikic
@msikic.bsky.social
· Mar 2
Reposted by Mile Sikic
Reposted by Mile Sikic
Reposted by Mile Sikic
Dmitry Antipov
@dantipov.bsky.social
· Dec 27

Verkko2: Integrating proximity ligation data with long-read De Bruijn graphs for efficient telomere-to-telomere genome assembly, phasing, and scaffolding
The Telomere-to-Telomere Consortium recently finished the first truly complete sequence of a human genome. To resolve the most complex repeats, this project relied on the semi-manual combination of lo...
www.biorxiv.org
Mile Sikic
@msikic.bsky.social
· Dec 22
Mile Sikic
@msikic.bsky.social
· Dec 22
Reposted by Mile Sikic
Javier Santoyo
@jsantoyo.bsky.social
· Dec 19
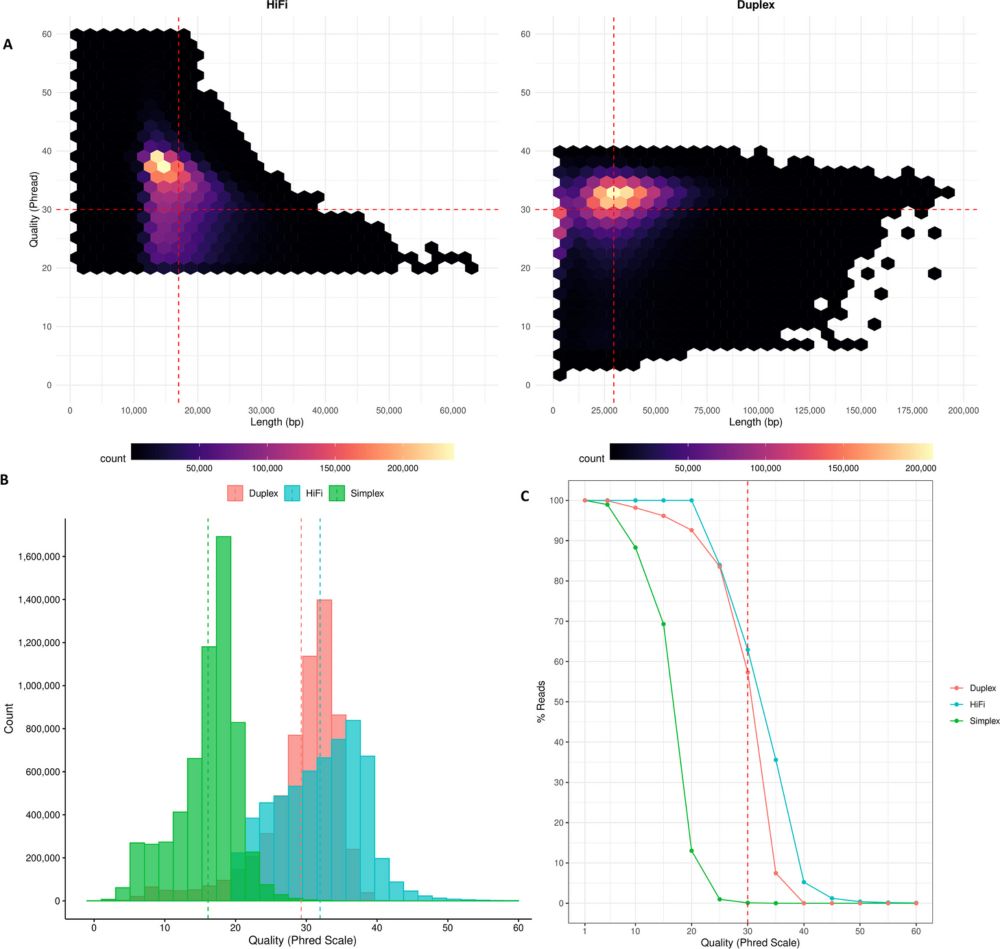
Evaluating data requirements for high-quality haplotype-resolved genomes for creating robust pangenome references - Genome Biology
Background Long-read technologies from Pacific Biosciences (PacBio) and Oxford Nanopore Technologies (ONT) have transformed genomics research by providing diverse data types like HiFi, Duplex, and ult...
genomebiology.biomedcentral.com
Mile Sikic
@msikic.bsky.social
· Dec 19
Mile Sikic
@msikic.bsky.social
· Dec 19

Evaluating data requirements for high-quality haplotype-resolved genomes for creating robust pangenome references - Genome Biology
Background Long-read technologies from Pacific Biosciences (PacBio) and Oxford Nanopore Technologies (ONT) have transformed genomics research by providing diverse data types like HiFi, Duplex, and ult...
genomebiology.biomedcentral.com
Mile Sikic
@msikic.bsky.social
· Dec 14

Detecting a wide range of epitranscriptomic modifications using a nanopore-sequencing-based computational approach with 1D score-clustering
Abstract. To date, over 40 epigenetic and 300 epitranscriptomic modifications have been identified. However, current short-read sequencing-based experiment
academic.oup.com