Nicola Bordin
@nbordin.bsky.social
840 followers
350 following
46 posts
Research Fellow in Bioinformatics (Proteins + ML + Function) @ CATH University College London | Mountains, Proteins, Food in that order | MSCA Alumn | Replies to emails!
Posts
Media
Videos
Starter Packs
Pinned
Nicola Bordin
@nbordin.bsky.social
· Nov 16
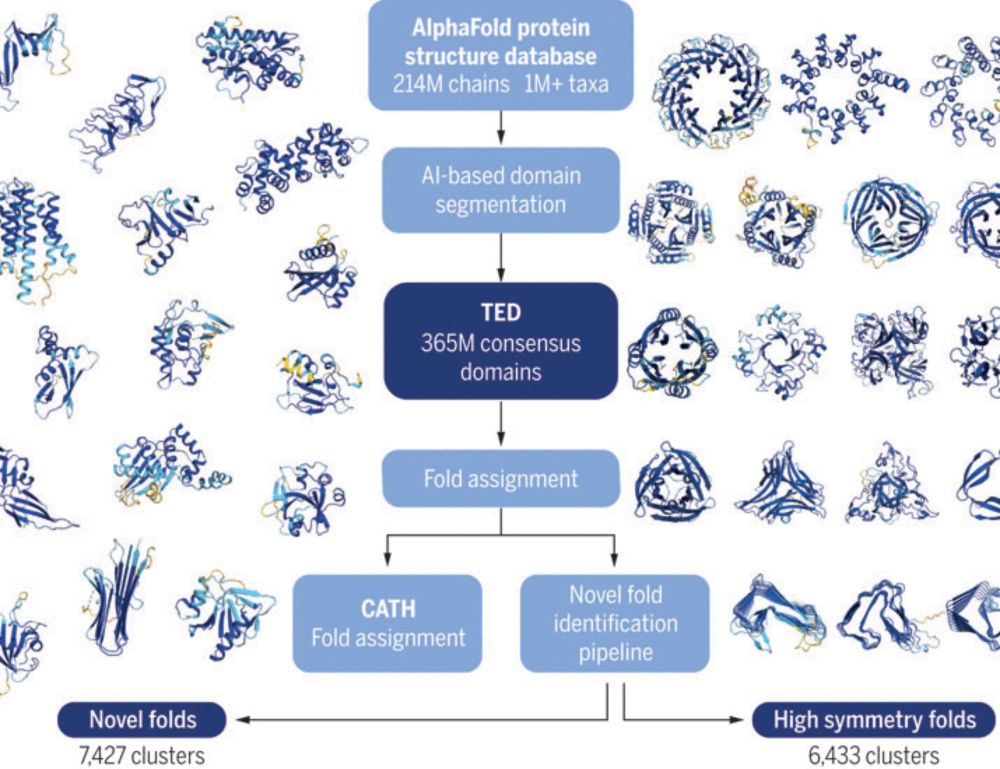
Exploring structural diversity across the protein universe with The Encyclopedia of Domains
The AlphaFold Protein Structure Database (AFDB) contains more than 214 million predicted protein structures composed of domains, which are independently folding units found in multiple structural and ...
www.science.org
Reposted by Nicola Bordin
Nicola Bordin
@nbordin.bsky.social
· Aug 26
Reposted by Nicola Bordin
CATH-Gene3D
@cathgene3d.bsky.social
· Aug 22
Reposted by Nicola Bordin
CATH-Gene3D
@cathgene3d.bsky.social
· Aug 22
Nicola Bordin
@nbordin.bsky.social
· Aug 13
Nicola Bordin
@nbordin.bsky.social
· Jul 20
Nicola Bordin
@nbordin.bsky.social
· Jul 12
Reposted by Nicola Bordin
Reposted by Nicola Bordin
Nicola Bordin
@nbordin.bsky.social
· Apr 29
Nicola Bordin
@nbordin.bsky.social
· Apr 29
Nicola Bordin
@nbordin.bsky.social
· Apr 28
Nicola Bordin
@nbordin.bsky.social
· Apr 28
Nicola Bordin
@nbordin.bsky.social
· Apr 28

Metagenomic-scale analysis of the predicted protein structure universe
Protein structure prediction breakthroughs, notably AlphaFold2 and ESMfold, have led to an unprecedented influx of computationally derived structures. The AlphaFold Protein Structure Database now prov...
www.biorxiv.org
Nicola Bordin
@nbordin.bsky.social
· Apr 9