Niko Klink
@nikolasklink.bsky.social
360 followers
330 following
24 posts
Chemical Biology & drug discovery @Max Planck Dortmund | All things Ubiquitin and bifunctionals @Gersch Lab.
Posts
Media
Videos
Starter Packs
Niko Klink
@nikolasklink.bsky.social
· Aug 11
Niko Klink
@nikolasklink.bsky.social
· Jul 10
Niko Klink
@nikolasklink.bsky.social
· Jul 9
Niko Klink
@nikolasklink.bsky.social
· Jul 6
Niko Klink
@nikolasklink.bsky.social
· Jul 6
Niko Klink
@nikolasklink.bsky.social
· Jul 6
Niko Klink
@nikolasklink.bsky.social
· Jul 6
Niko Klink
@nikolasklink.bsky.social
· Jul 5
Niko Klink
@nikolasklink.bsky.social
· Jul 5
Niko Klink
@nikolasklink.bsky.social
· Jul 5
Niko Klink
@nikolasklink.bsky.social
· Jul 5
Niko Klink
@nikolasklink.bsky.social
· Jul 5

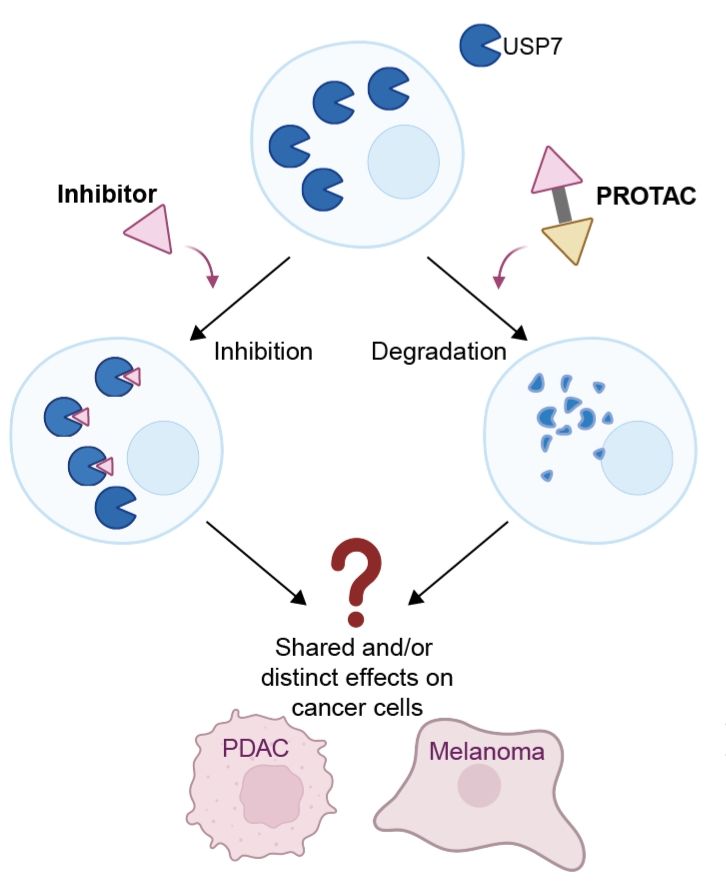
Targeted degradation of USP7 in solid cancer cells reveals disparate effects of deubiquitinase inhibition vs. acute protein depletion
Proteolysis-targeting chimeras (PROTACs) co-op the ubiquitin system for targeted protein degradation, creating opportunities to interrogate cellular functions of proteins through "chemical knockdown"....
www.biorxiv.org
Niko Klink
@nikolasklink.bsky.social
· May 5
Niko Klink
@nikolasklink.bsky.social
· May 5
Niko Klink
@nikolasklink.bsky.social
· May 5

Chimeric deubiquitinase engineering reveals structural basis for specific inhibition of the mitophagy regulator USP30 - Nature Structural & Molecular Biology
Kazi et al. report the crystal structure of the mitochondrial deubiquitinase USP, a clinical stage Parkinson’s disease drug target, in complex with a specific inhibitor. The authors delineate a framew...
www.nature.com
Niko Klink
@nikolasklink.bsky.social
· Mar 25
Reposted by Niko Klink
Cynthia Wolberger
@cwolberger.bsky.social
· Mar 21
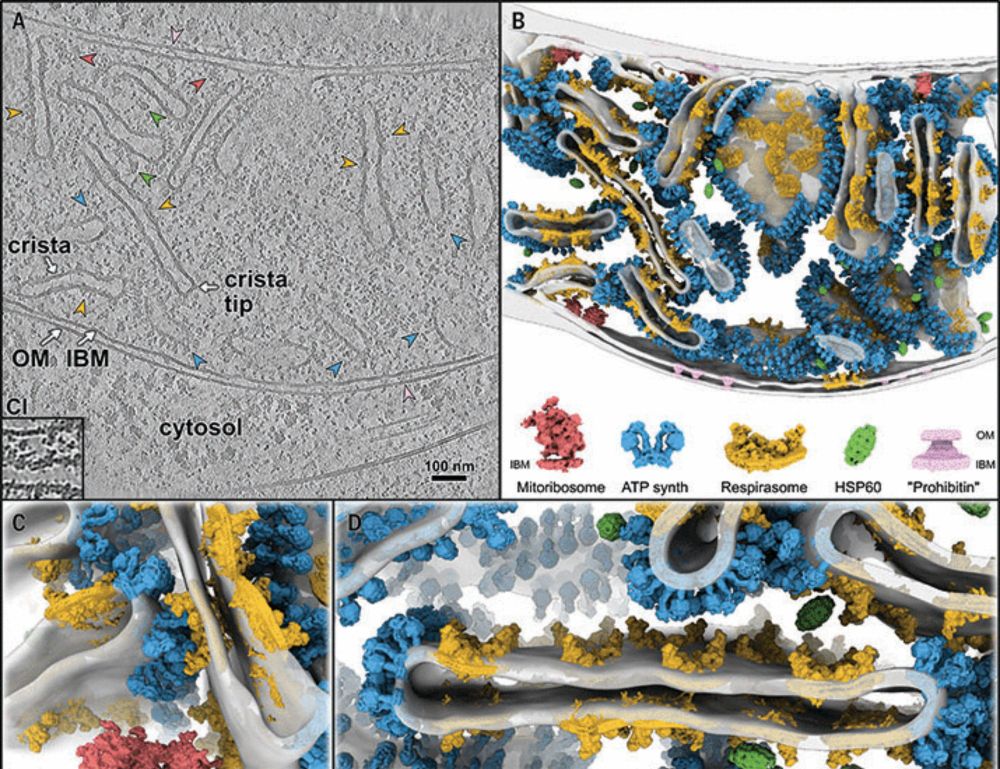
In-cell architecture of the mitochondrial respiratory chain
Mitochondria regenerate adenosine triphosphate (ATP) through oxidative phosphorylation. This process is carried out by five membrane-bound complexes collectively known as the respiratory chain, workin...
www.science.org
Reposted by Niko Klink
Mike Cohen
@michaelnadbio.bsky.social
· Feb 25

Ubiquitin is directly linked via an ester to protein-conjugated mono-ADP-ribose | The EMBO Journal
imageimageCertain E3 ligases have been found to ubiquitylate hydroxyl groups on free NAD+ and
ADP-ribose in vitro, but the in vivo occurrence of this dual post-translational modification
has remained ...
www.embopress.org
Niko Klink
@nikolasklink.bsky.social
· Dec 23