Nori Zolboot
@norjin.bsky.social
44 followers
77 following
18 posts
• Postdoc @scripps.edu Neuroscience
• MicroRNAs | Purkinje cells | RNA therapeutics
• What is sarcasm?
• PhD from Scripps Research ‘23
• BS from Nagoya University ‘17
• 🇲🇳->🇯🇵->🇺🇸 | 🔬🐱🧶🍫🎱🎬
Posts
Media
Videos
Starter Packs
Reposted by Nori Zolboot
Giordano Lippi
@lippilab.bsky.social
· May 21
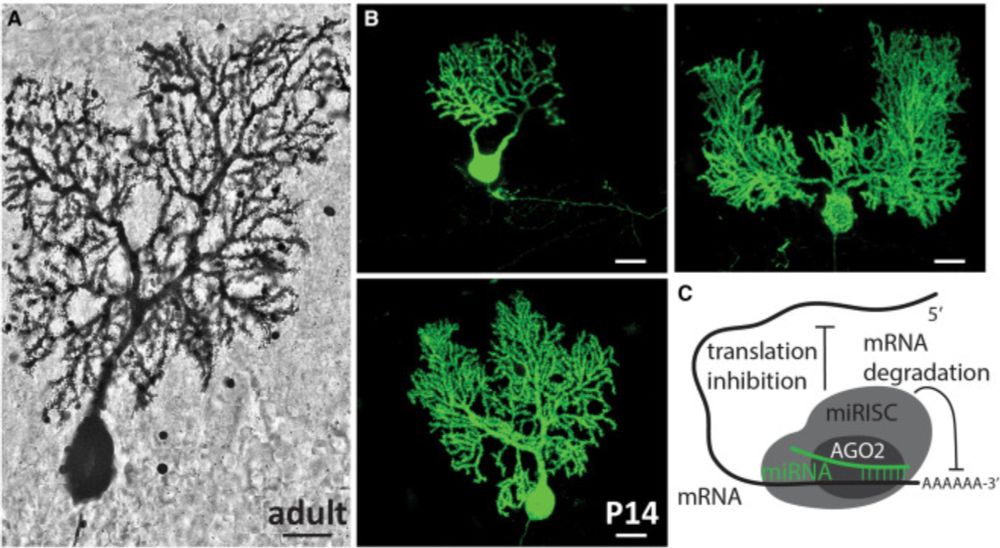
Rooted in regulation: MicroRNAs as gardeners of the Purkinje cell dendritic arbor
Zolboot et al.1 developed tools to manipulate and map microRNA function in cerebellar
Purkinje cells with temporal precision, revealing key roles in dendritic development
and circuit connectivity. The...
www.cell.com
Nori Zolboot
@norjin.bsky.social
· May 21
Reposted by Nori Zolboot
Reposted by Nori Zolboot
Neuron
@cp-neuron.bsky.social
· Apr 2

MicroRNA mechanisms instructing Purkinje cell specification
Zolboot et al. introduce DD-T6B and SAP-seq, methods for investigating microRNA function with enhanced spatiotemporal resolution, and identify Purkinje cell-specific microRNAs and targets instructing its unique dendritic arborization and climbing fiber synaptogenesis. Their findings highlight the critical role of post-transcriptional regulatory mechanisms and demonstrate the broad applicability of their tools.
dlvr.it
Nori Zolboot
@norjin.bsky.social
· Apr 2
Nori Zolboot
@norjin.bsky.social
· Apr 2
Nori Zolboot
@norjin.bsky.social
· Apr 2
Nori Zolboot
@norjin.bsky.social
· Apr 2
Nori Zolboot
@norjin.bsky.social
· Apr 2
Nori Zolboot
@norjin.bsky.social
· Apr 2
Nori Zolboot
@norjin.bsky.social
· Apr 2
Nori Zolboot
@norjin.bsky.social
· Apr 2
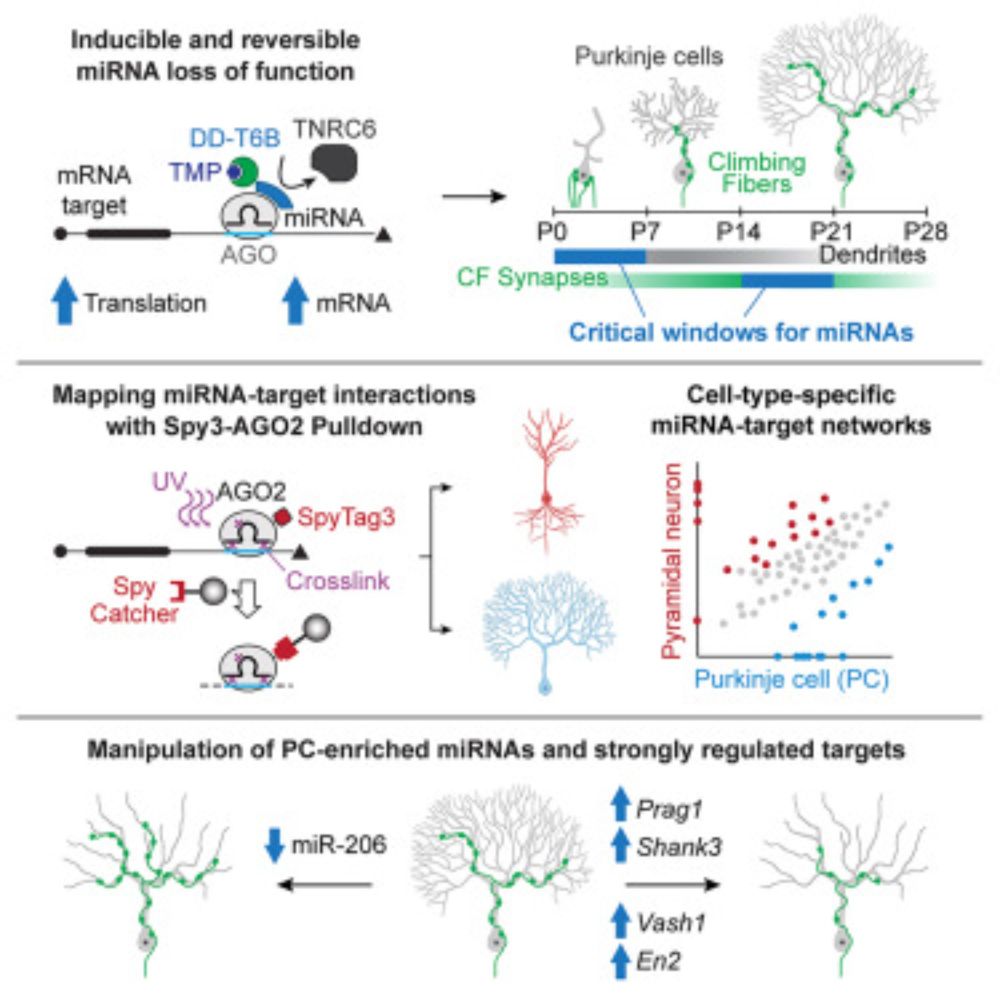
MicroRNA mechanisms instructing Purkinje cell specification
Zolboot et al. introduce DD-T6B and SAP-seq, methods for investigating microRNA function
with enhanced spatiotemporal resolution, and identify Purkinje cell-specific microRNAs
and targets instructing ...
www.cell.com
Reposted by Nori Zolboot
Reposted by Nori Zolboot
Nature Neuroscience
@natneuro.nature.com
· Dec 18

Stepwise molecular specification of excitatory synapse diversity onto cerebellar Purkinje cells - Nature Neuroscience
Brain function requires the formation of diverse and specific synapses. The authors show that the molecular code specifying excitatory connectivity on Purkinje cells evolves in an input-specific manne...
www.nature.com
Nori Zolboot
@norjin.bsky.social
· Nov 19