Ophir Shalem
@ophirshalem.bsky.social
1.3K followers
740 following
13 posts
Scientist, Associate Professor, Department of Genetics, Perelman School of Medicine, University of Pennsylvania, Children’s Hospital of Philadelphia
Functional genomics, Proteostasis, Neurodegeneration
www.shalemlab.org
Posts
Media
Videos
Starter Packs
Pinned
Ophir Shalem
@ophirshalem.bsky.social
· Apr 23
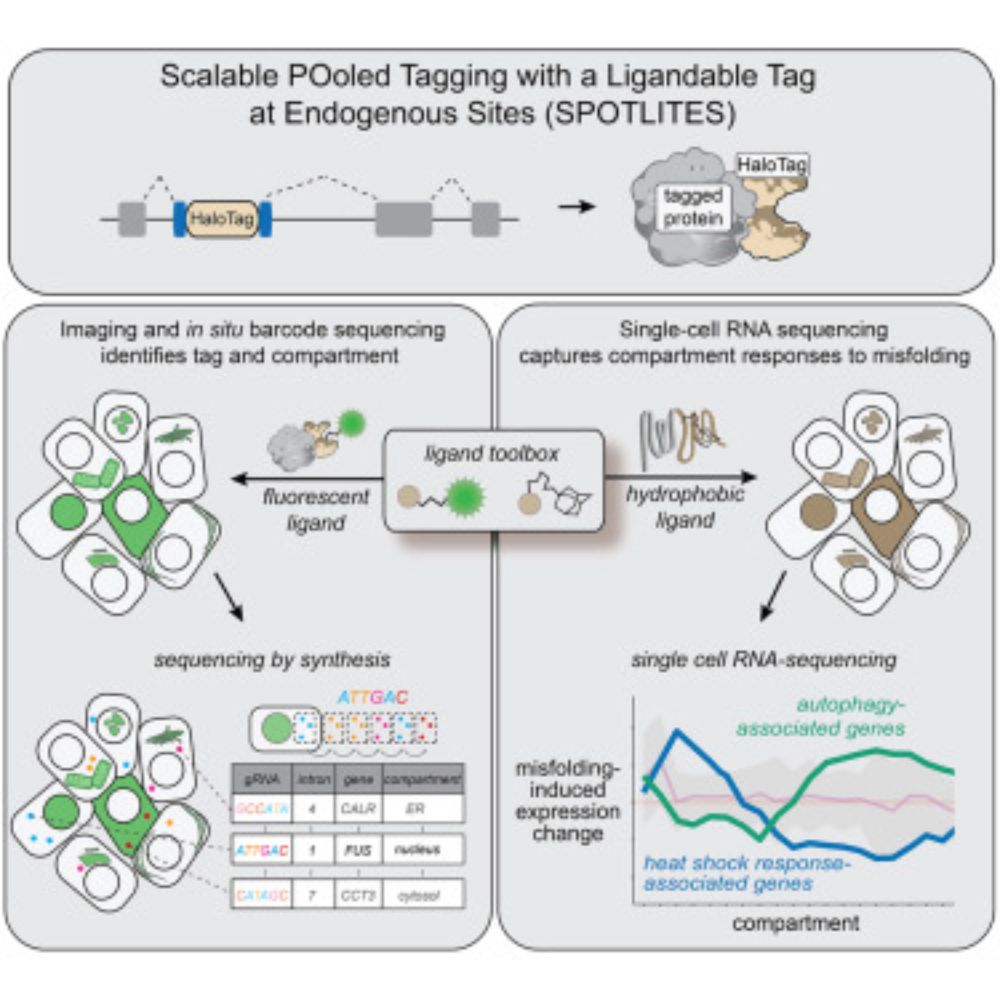
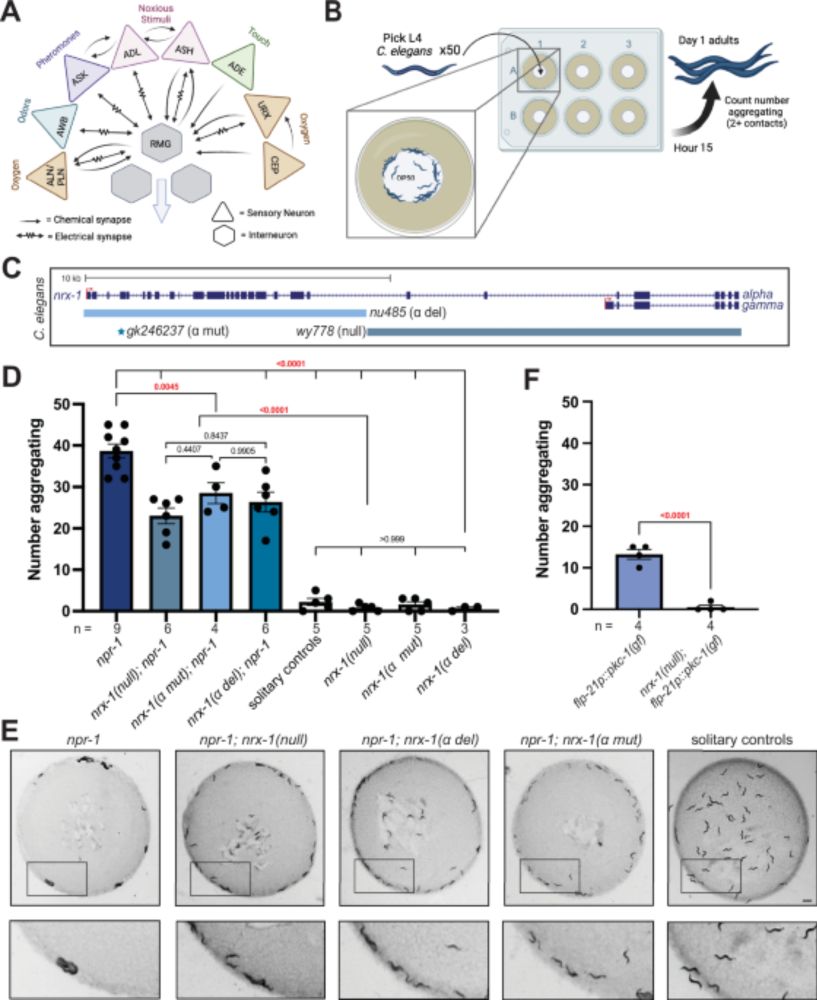
Pooled tagging and hydrophobic targeting of endogenous proteins for unbiased mapping of unfolded protein responses
Sansbury and Serebrenik et al. use pooled gene tagging with in situ sequencing and
high-throughput image analysis to generate a cell pool with endogenous HaloTag fusions.
Hydrophobic targeting is then...
www.cell.com
Reposted by Ophir Shalem
John Murray
@jisaacmurray.bsky.social
· Jun 20
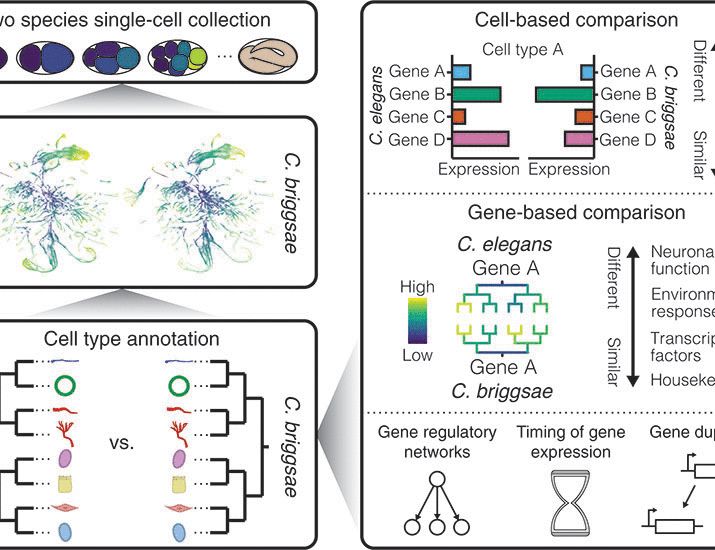
Lineage-resolved analysis of embryonic gene expression evolution in C. elegans and C. briggsae
The constraints that govern the evolution of gene expression patterns across development remain unclear. Single-cell RNA sequencing can detail these constraints by systematically profiling homologous ...
www.science.org
Ophir Shalem
@ophirshalem.bsky.social
· Jun 17
Ophir Shalem
@ophirshalem.bsky.social
· Apr 23

Pooled tagging and hydrophobic targeting of endogenous proteins for unbiased mapping of unfolded protein responses
Sansbury and Serebrenik et al. use pooled gene tagging with in situ sequencing and
high-throughput image analysis to generate a cell pool with endogenous HaloTag fusions.
Hydrophobic targeting is then...
www.cell.com
Reposted by Ophir Shalem
Reposted by Ophir Shalem
George Burslem
@gburslem.bsky.social
· Feb 28

Class I histone deacetylases catalyze lysine lactylation
Metabolism and post-translational modifications (PTMs) are intrinsically linked and the number of identified metabolites that can covalently modify proteins continues to increase. This metabolism/PTM ...
www.biorxiv.org
Reposted by Ophir Shalem
Reposted by Ophir Shalem
Doug Epstein
@epsteind.bsky.social
· Nov 22
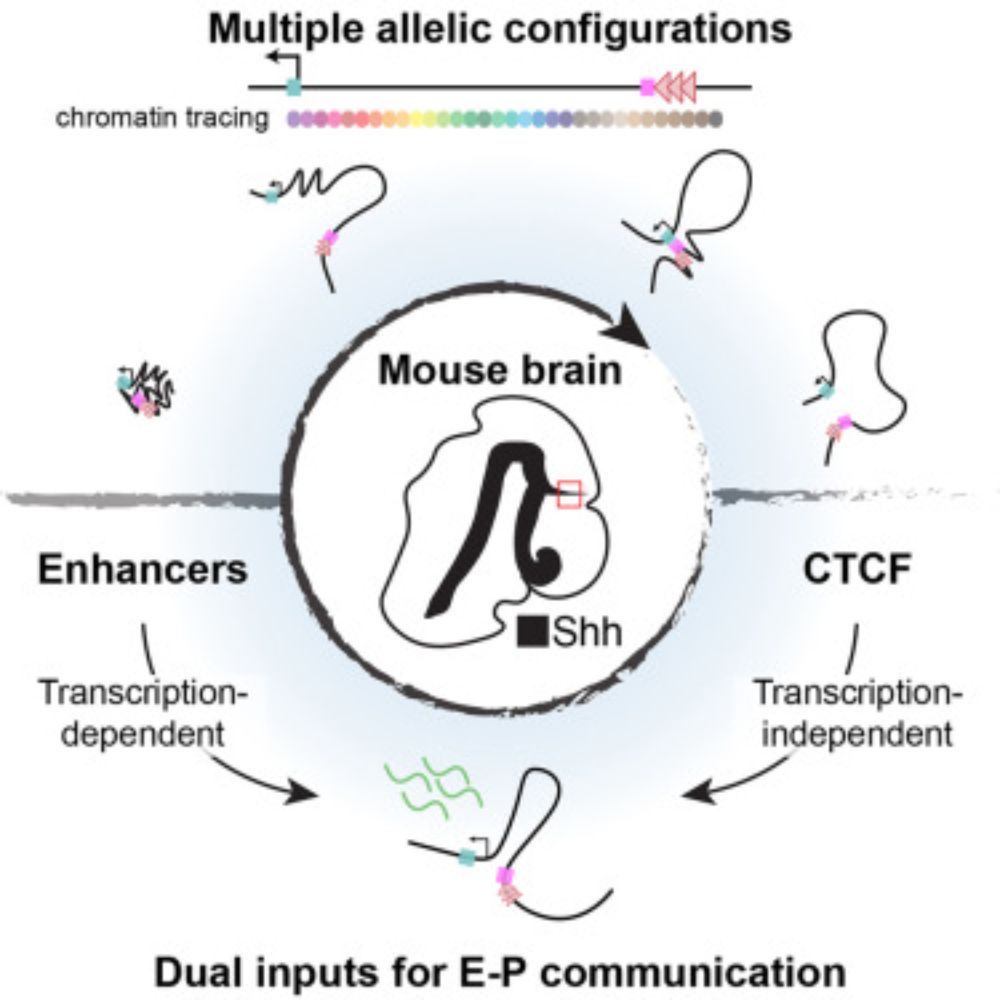
Multiple allelic configurations govern long-range Shh enhancer-promoter communication in the embryonic forebrain
Harke et al. use sequential DNA-FISH to achieve single-allele resolution of the Sonic
hedgehog (Shh) regulatory domain in the mouse forebrain. They show that multiple allelic
configurations occur inde...
www.cell.com
Reposted by Ophir Shalem
Struan Grant
@struangrant.bsky.social
· Nov 25

Recruitment Summit for Postdoctoral Scholars in Cell Biology and Genomics | Children’s Hospital of Philadelphia
Recruitment Summit for Postdoctoral Scholars in Cell Biology and Genomics in Philadelphia, Pennsylvania, United States. null at Children’s Hospital of Philadelphia
careers.chop.edu
Reposted by Ophir Shalem
John Doench
@johndoench.bsky.social
· Nov 15

Activity-based selection for enhanced base editor mutational scanning
Base editing is a CRISPR-based technology that enables high-throughput, nucleotide-level functional interrogation of the genome, which is essential for understanding the genetic basis of human disease...
www.biorxiv.org
Reposted by Ophir Shalem
Michael Ward
@michael-e-ward.bsky.social
· Nov 15
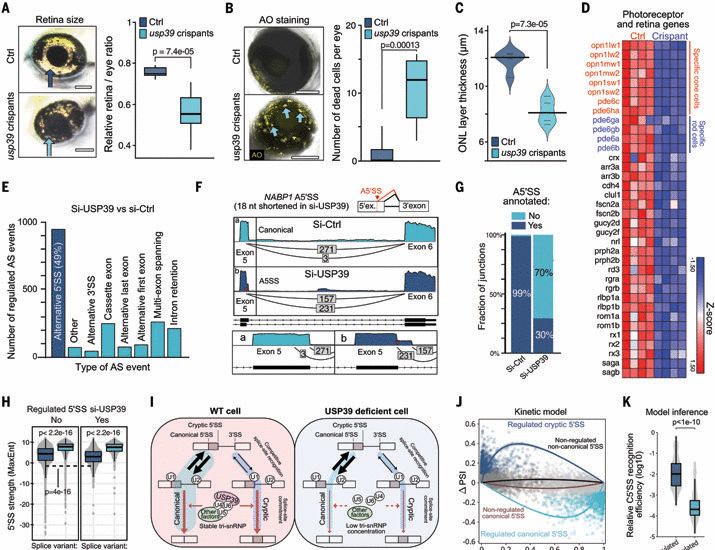
Pathogenic proteotoxicity of cryptic splicing is alleviated by ubiquitination and ER-phagy
RNA splicing enables the functional adaptation of cells to changing contexts. Impaired splicing has been associated with diseases, including retinitis pigmentosa, but the underlying molecular mechanis...
www.science.org
Ophir Shalem
@ophirshalem.bsky.social
· Nov 10
Ophir Shalem
@ophirshalem.bsky.social
· Nov 10
Ophir Shalem
@ophirshalem.bsky.social
· Nov 10
Ophir Shalem
@ophirshalem.bsky.social
· Nov 10
Ophir Shalem
@ophirshalem.bsky.social
· Nov 10
Ophir Shalem
@ophirshalem.bsky.social
· Nov 10
Ophir Shalem
@ophirshalem.bsky.social
· Nov 10

CRISPR associated enzymes are mislocalized to the cytoplasm in iPSC-derived neurons resulting in KRAB-specific degradation
The use of CRISPR-associated enzymes in iPSC-derived neurons for precise gene targeting and high-throughput gene perturbation screens offers great potential but presents unique challenges compared to ...
www.biorxiv.org