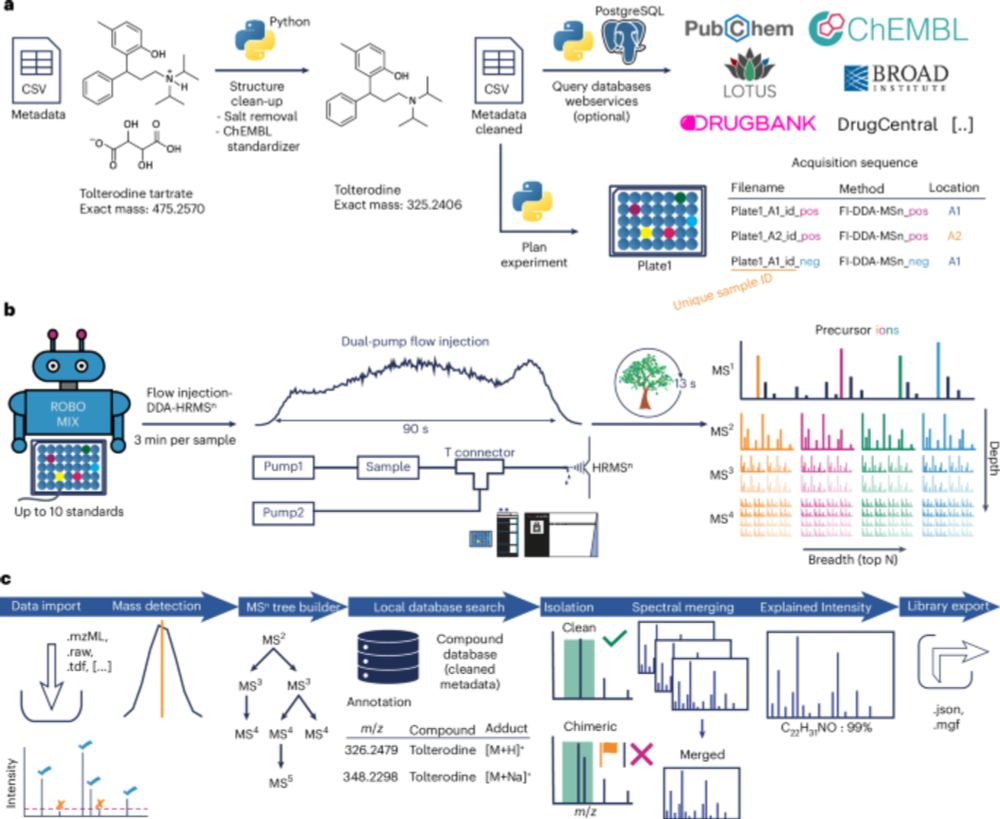
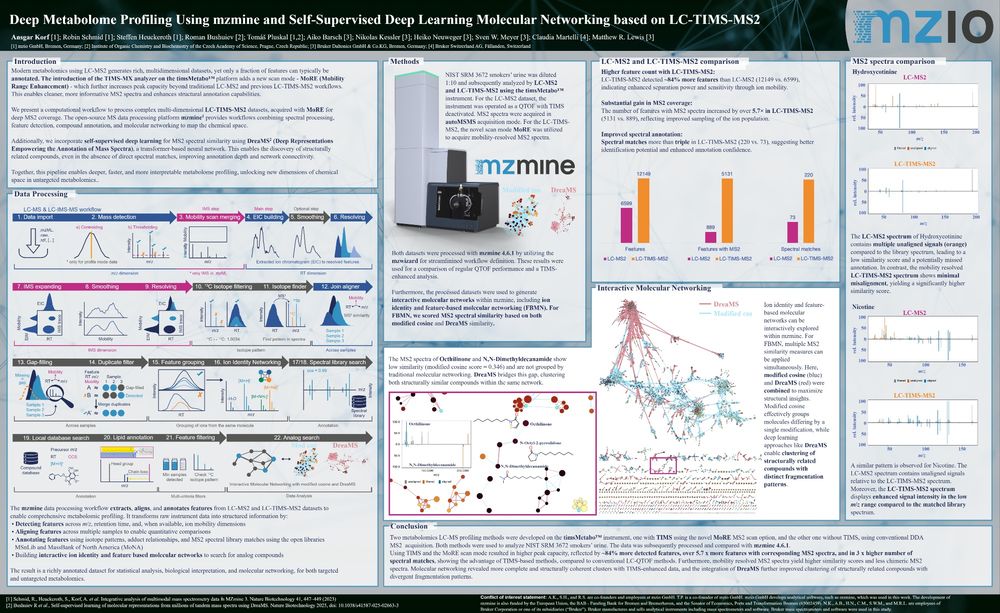
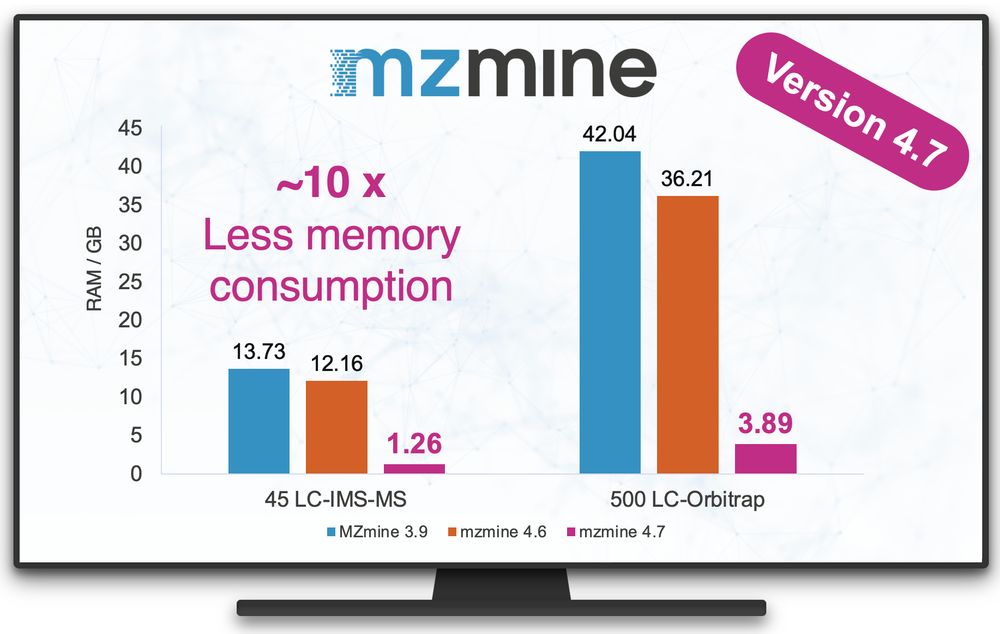
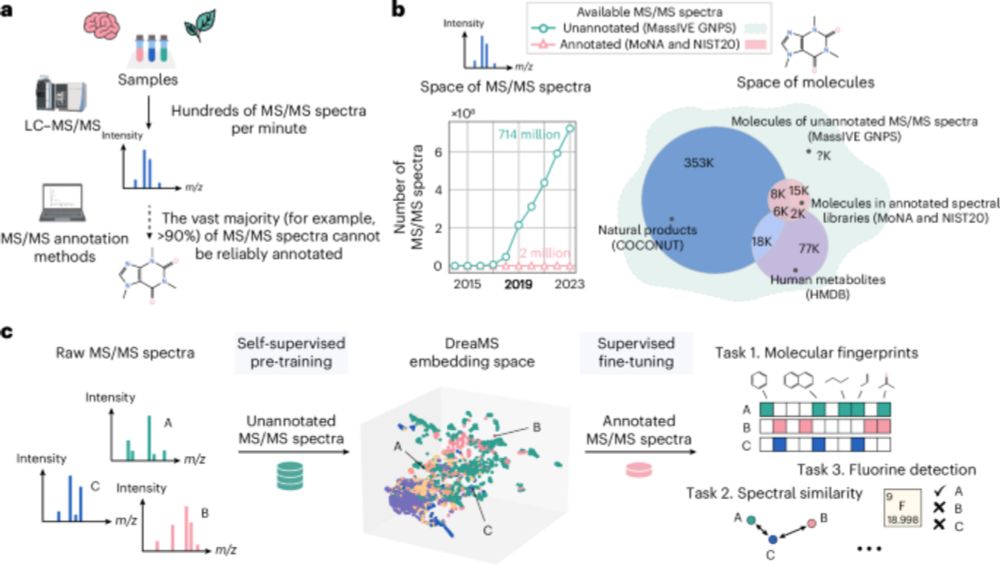
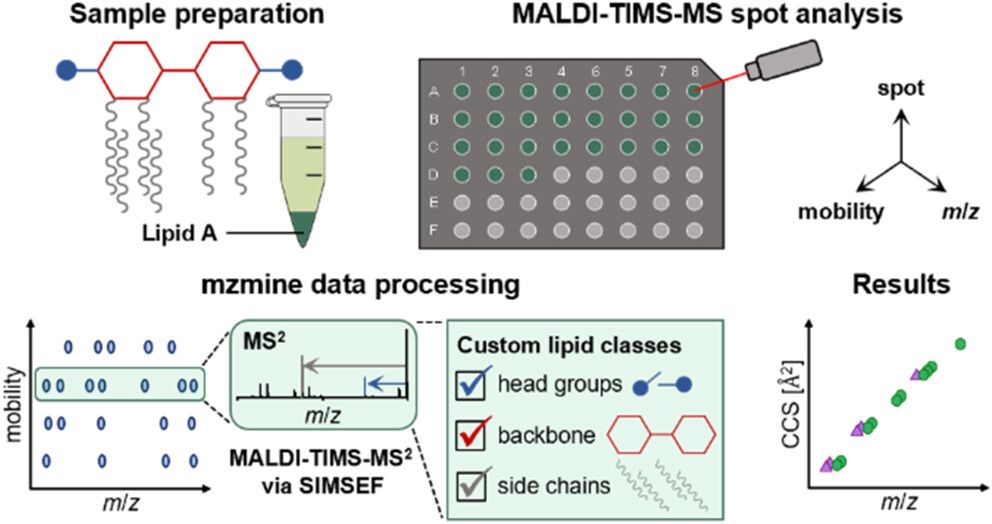
Tomáš Pluskal
@pluskal-lab.org
540 followers
490 following
17 posts
Group Leader at the Institute of Organic Chemistry and Biochemistry (IOCB Prague @iocbprague.bsky.social). Plant biochemistry, metabolomics, genomics, bioML, etc. https://www.pluskal-lab.org
Posts
Media
Videos
Starter Packs
Reposted by Tomáš Pluskal
Tomáš Pluskal
@pluskal-lab.org
· Jun 27
Reposted by Tomáš Pluskal
Reposted by Tomáš Pluskal
Reposted by Tomáš Pluskal
Reposted by Tomáš Pluskal
Reposted by Tomáš Pluskal
Reposted by Tomáš Pluskal
Reposted by Tomáš Pluskal
Reposted by Tomáš Pluskal
Tomáš Pluskal
@pluskal-lab.org
· Apr 29
Reposted by Tomáš Pluskal
Reposted by Tomáš Pluskal
Tomáš Pluskal
@pluskal-lab.org
· Mar 11