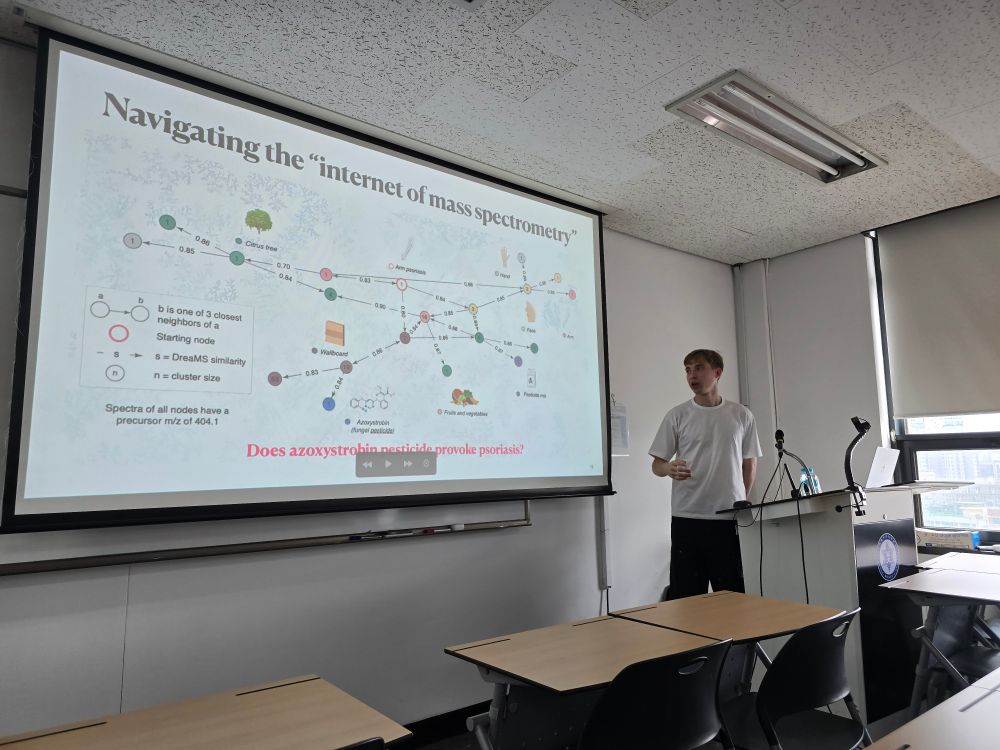
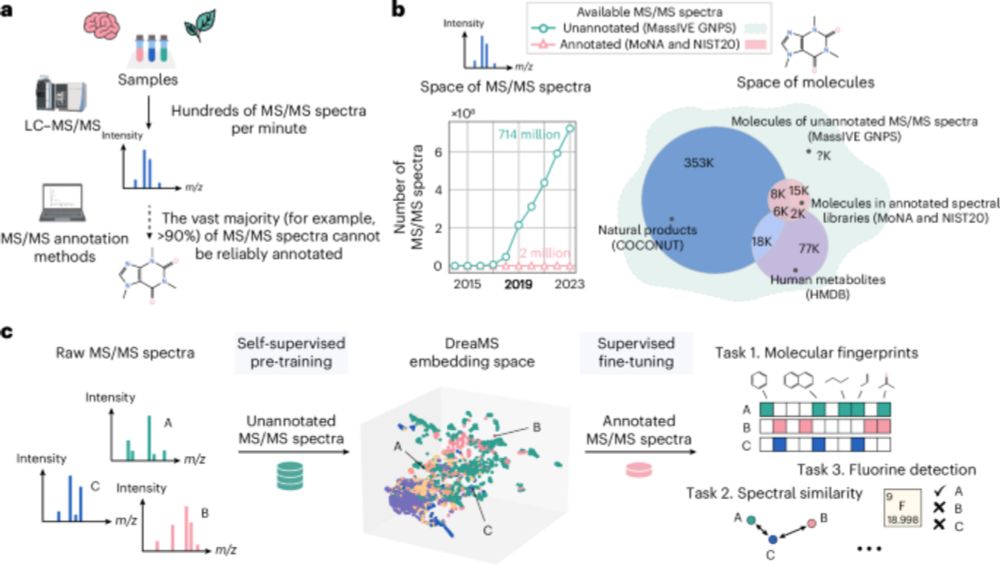
Roman Bushuiev
@roman-bushuiev.bsky.social
180 followers
220 following
19 posts
🌏 https://roman-bushuiev.github.io/
Posts
Media
Videos
Starter Packs
Reposted by Roman Bushuiev
Reposted by Roman Bushuiev
Reposted by Roman Bushuiev
Reposted by Roman Bushuiev
Reposted by Roman Bushuiev
Reposted by Roman Bushuiev
Reposted by Roman Bushuiev