Patricia Nano
@prnano9.bsky.social
59 followers
91 following
15 posts
Postdoc @bhadurilab.bsky.social
Human brain development + single-cell omics
Posts
Media
Videos
Starter Packs
Pinned
Patricia Nano
@prnano9.bsky.social
· May 2
Nature Neuroscience
@natneuro.nature.com
· Apr 29

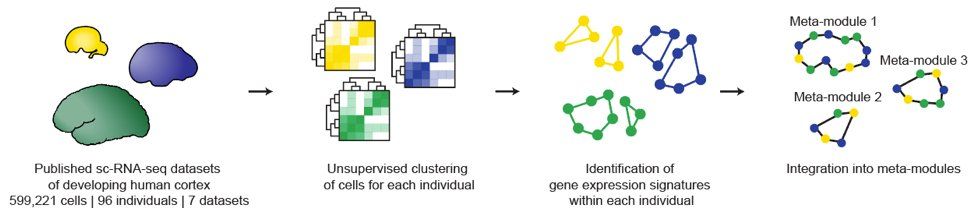
Integrated analysis of molecular atlases unveils modules driving developmental cell subtype specification in the human cortex - Nature Neuroscience
Nano et al. introduce a pipeline to generate meta-atlases of the human brain from existing single-cell datasets and extract gene modules linked to cell fate specification. Perturbing these programs in...
www.nature.com
Reposted by Patricia Nano
Reposted by Patricia Nano
Patricia Nano
@prnano9.bsky.social
· May 2
Patricia Nano
@prnano9.bsky.social
· May 2
Patricia Nano
@prnano9.bsky.social
· May 2
Patricia Nano
@prnano9.bsky.social
· May 2
Patricia Nano
@prnano9.bsky.social
· May 2
Patricia Nano
@prnano9.bsky.social
· May 2
Patricia Nano
@prnano9.bsky.social
· May 2
Patricia Nano
@prnano9.bsky.social
· May 2
Patricia Nano
@prnano9.bsky.social
· May 2
Patricia Nano
@prnano9.bsky.social
· May 2
Nature Neuroscience
@natneuro.nature.com
· Apr 29

Integrated analysis of molecular atlases unveils modules driving developmental cell subtype specification in the human cortex - Nature Neuroscience
Nano et al. introduce a pipeline to generate meta-atlases of the human brain from existing single-cell datasets and extract gene modules linked to cell fate specification. Perturbing these programs in...
www.nature.com