Ramon Massoni Badosa
@rmassonix.bsky.social
110 followers
170 following
16 posts
Postdoc at NYGC and WCM in love with single-cell multiomics, immunology, hematology, data science. I care about open science and education
Posts
Media
Videos
Starter Packs
Reposted by Ramon Massoni Badosa
Reposted by Ramon Massoni Badosa
Reposted by Ramon Massoni Badosa
Anshul Kundaje
@anshulkundaje.bsky.social
· Jan 24
Reposted by Ramon Massoni Badosa
Anshul Kundaje
@anshulkundaje.bsky.social
· Jan 13
Reposted by Ramon Massoni Badosa
Benoit Bruneau
@benoitbruneau.bsky.social
· Jan 13
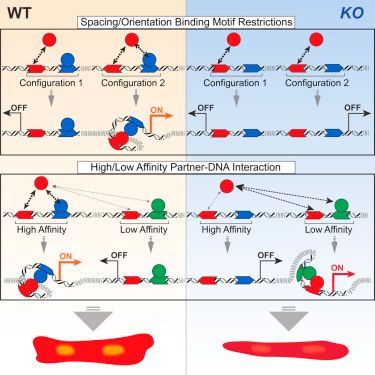
Complex Interdependence Regulates Heterotypic Transcription Factor Distribution and Coordinates Cardiogenesis
Transcription factors (TFs) are thought to function with partners to achieve specificity and precise quantitative outputs. In the developing heart, he…
www.sciencedirect.com
Pau Badia i Mompel
@paubadiam.bsky.social
· Dec 23
Saez-Rodriguez Group
@saezlab.bsky.social
· Dec 23
Reposted by Ramon Massoni Badosa
Anshul Kundaje
@anshulkundaje.bsky.social
· Dec 25
Reposted by Ramon Massoni Badosa
Prachee Avasthi
@pracheeac.bsky.social
· Dec 22
Richard Sever
@richardsever.bsky.social
· Dec 21