Rowan Green
@rowancallumg.bsky.social
560 followers
1.1K following
36 posts
Postdoc studying viral evolution with @asherleeks.bsky.social | previously @mermanchester.bsky.social | mutagenesis | bacteria | viruses | big R nerd 🧬🦠💻
🏳️⚧️ He/Him
Posts
Media
Videos
Starter Packs
Rowan Green
@rowancallumg.bsky.social
· Aug 18

Spontaneous mutation rate is a plastic trait associated with population density across domains of life
Author summary Spontaneous mutations fuel evolution, but the rate at which they occur can vary for a particular organism depending on its environment—a phenomenon known as mutation-rate plasticity. Fo...
doi.org
Reposted by Rowan Green
Zamin Iqbal
@zaminiqbal.bsky.social
· Aug 18
Rowan Green
@rowancallumg.bsky.social
· Aug 18

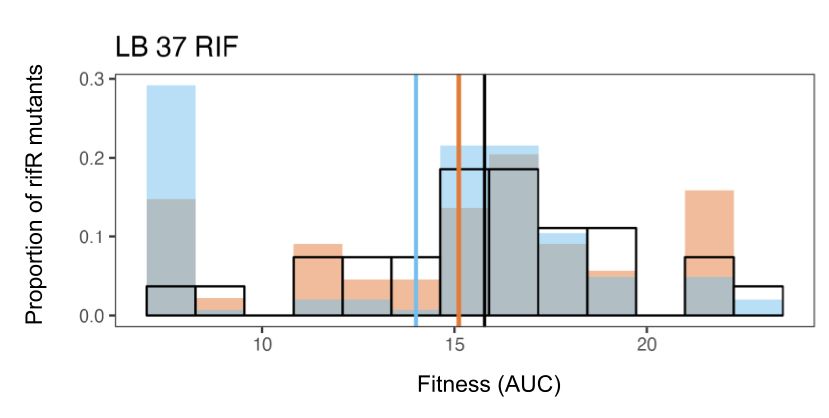
Antimutator and Mutational Spectrum Effects Can Combine to Reduce Evolutionary Potential in Escherichia coli ΔnudJ
Abstract. The rate of spontaneous mutation is a key factor in determining the capacity of a population to adapt to a novel environment, for example, a bact
doi.org
Rowan Green
@rowancallumg.bsky.social
· Aug 18
Rowan Green
@rowancallumg.bsky.social
· Aug 18
Rowan Green
@rowancallumg.bsky.social
· Aug 18
Rowan Green
@rowancallumg.bsky.social
· Aug 18
Rowan Green
@rowancallumg.bsky.social
· Aug 18
Rowan Green
@rowancallumg.bsky.social
· Aug 18
Rowan Green
@rowancallumg.bsky.social
· Aug 18
Rowan Green
@rowancallumg.bsky.social
· Aug 18

Antimutator and Mutational Spectrum Effects Can Combine to Reduce Evolutionary Potential in Escherichia coli ΔnudJ
Abstract. The rate of spontaneous mutation is a key factor in determining the capacity of a population to adapt to a novel environment, for example, a bact
doi.org
Reposted by Rowan Green
Reposted by Rowan Green
Rowan Green
@rowancallumg.bsky.social
· May 27
Reposted by Rowan Green
Bruno Luviano
@brunoluviano.bsky.social
· May 15
Reposted by Rowan Green
Peter Lind
@peteralind.bsky.social
· Mar 13

Postdoctor, 2 years, in predicting the evolution of antibiotic resistance
The Department of Molecular Biology seeks a postdoctoral researcher who will work with forecasting and steering experimental evolution of antibiotic resistance. The employment is full-time for two yea
umu.varbi.com
Reposted by Rowan Green
Rui Borges
@ruiborges23.bsky.social
· Jan 27

The rarity of mutations and the inflation of bacterial effective population sizes
Mutations are fundamental for evolution, and their mathematical modelling in population genetics heavily relies on our perception of their frequency and the timescale over which they occur. A comm...
besjournals.onlinelibrary.wiley.com