Samuel Maiwald
@samuelmaiwald.bsky.social
140 followers
360 following
18 posts
BIF PhD student in Schulman lab @mpibiochem.bsky.social, interested in structural biology and ubiquitin system
Posts
Media
Videos
Starter Packs
Pinned
Samuel Maiwald
@samuelmaiwald.bsky.social
· May 26
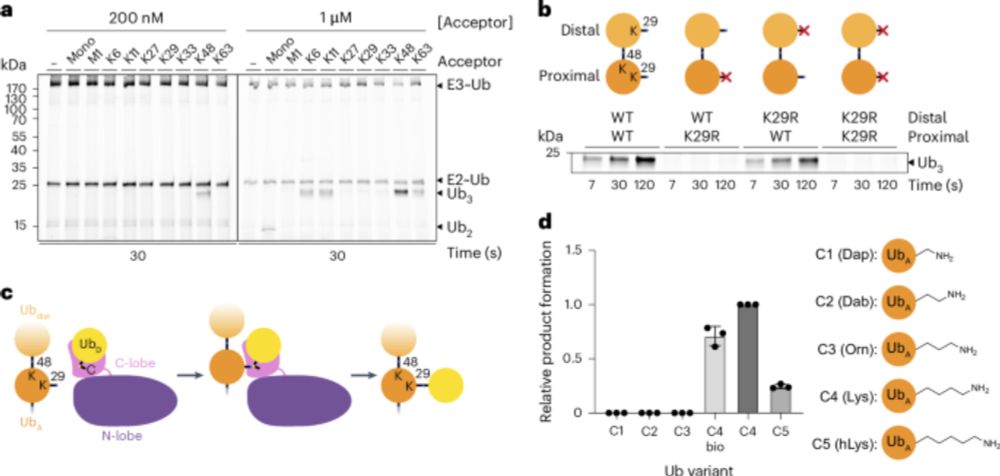
TRIP12 structures reveal HECT E3 formation of K29 linkages and branched ubiquitin chains - Nature Structural & Molecular Biology
Using biochemistry, chemical biology, and cryo-EM, Maiwald et al. elucidate how TRIP12 forms K29 linkages and K29/K48-linked branched ubiquitin chains, revealing a mechanism for polyubiquitylation sha...
www.nature.com
Reposted by Samuel Maiwald
Carolin Klose
@carolinklose.bsky.social
· Aug 11

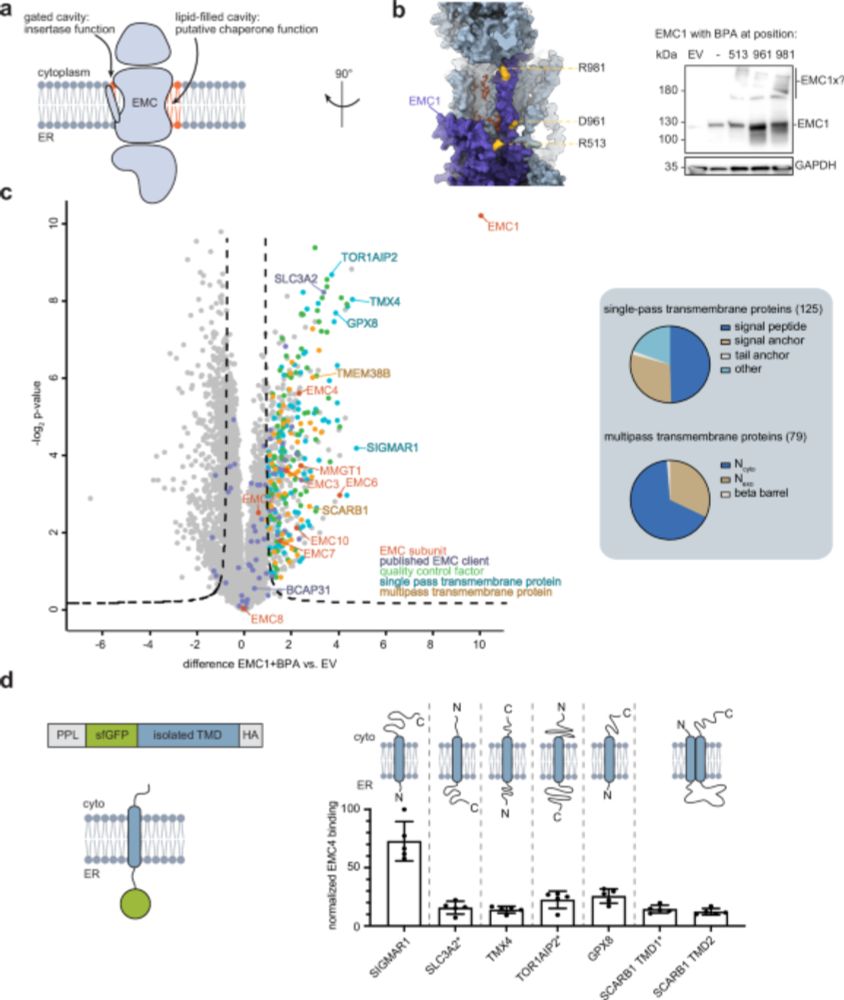
Structural basis of an EMC:Spf1 insertase-dislocase complex in the eukaryotic endoplasmic reticulum
Most eukaryotic membrane proteins are inserted into the membrane at the endoplasmic reticulum (ER). This essential but error-prone process relies on molecular quality control machineries to prevent mi...
www.biorxiv.org
Reposted by Samuel Maiwald
Samuel Maiwald
@samuelmaiwald.bsky.social
· May 26
Reposted by Samuel Maiwald
Samuel Maiwald
@samuelmaiwald.bsky.social
· May 26
Samuel Maiwald
@samuelmaiwald.bsky.social
· May 26
Samuel Maiwald
@samuelmaiwald.bsky.social
· May 26
Samuel Maiwald
@samuelmaiwald.bsky.social
· May 26
Samuel Maiwald
@samuelmaiwald.bsky.social
· May 26

TRIP12 structures reveal HECT E3 formation of K29 linkages and branched ubiquitin chains - Nature Structural & Molecular Biology
Using biochemistry, chemical biology, and cryo-EM, Maiwald et al. elucidate how TRIP12 forms K29 linkages and K29/K48-linked branched ubiquitin chains, revealing a mechanism for polyubiquitylation sha...
www.nature.com
Samuel Maiwald
@samuelmaiwald.bsky.social
· May 26
Samuel Maiwald
@samuelmaiwald.bsky.social
· May 26
Reposted by Samuel Maiwald
Leo Kiss
@leokiss.bsky.social
· Mar 24
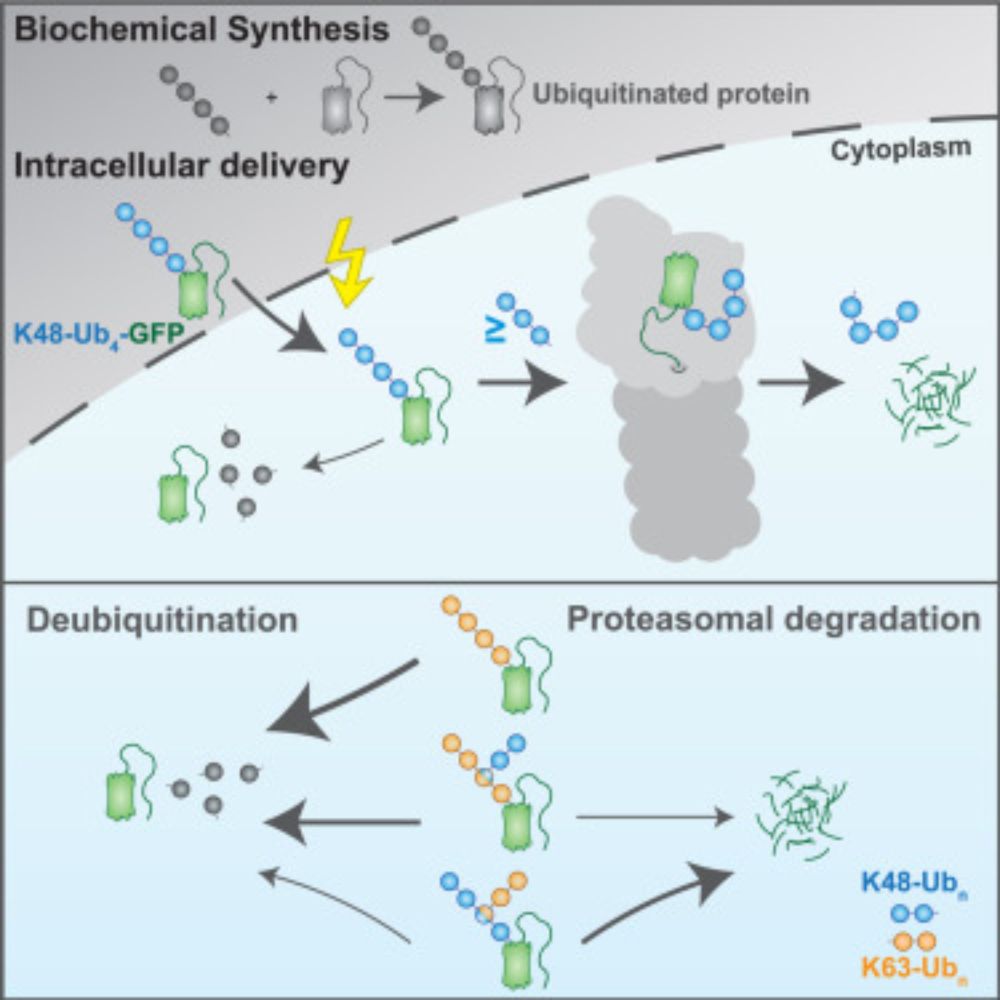
UbiREAD deciphers proteasomal degradation code of homotypic and branched K48 and K63 ubiquitin chains
Ubiquitin chains determine the fates of their modified proteins, including proteasomal
degradation. Kiss et al. present UbiREAD, a technology to monitor cellular degradation
and deubiquitination at hi...
www.cell.com