Sarah Gurev
@sarahgurev.bsky.social
89 followers
190 following
13 posts
Postdoc @ Debbie Marks Lab, Harvard | Prev. PhD @ MIT EECS || ML for Proteins + Viruses 🦠
Posts
Media
Videos
Starter Packs
Reposted by Sarah Gurev
Reposted by Sarah Gurev
Bloom lab
@jbloomlab.bsky.social
· Sep 8

Near real-time data on the human neutralizing antibody landscape to influenza virus to inform vaccine-strain selection in September 2025
The hemagglutinin of human influenza virus evolves rapidly to erode neutralizing antibody immunity. Twice per year, new vaccine strains are selected with the goal of providing maximum protection again...
www.biorxiv.org
Reposted by Sarah Gurev
Jon Mifsud
@jonathonmifsud.bsky.social
· Aug 25

Recent advances in the inference of deep viral evolutionary history | Journal of Virology
Phylogenetic studies examining the origins, emergence, and spread of viruses have arguably been one of the most active and successful areas of evolutionary biology and form the bedrock of the flourishing field of genomic epidemiology. This, in part, reflects the ability of viruses, particularly those with RNA genomes, to evolve at rates much greater than their cellular counterparts (1). The rapid rate at which viruses evolve and accumulate mutations enables evolutionary signals to be identified through comparative genomics at short timescales relevant for outbreak investigation and response. The integration of phylogenetics and epidemiology, known as phylodynamics, has become a vital tool in response to numerous viral outbreaks, epidemics, and pandemics, including Ebola (2), Zika (3), and, more recently, COVID-19 (4) and mpox (5).
journals.asm.org
Reposted by Sarah Gurev
Erin Doherty
@erinedoherty.bsky.social
· Aug 22

Divergent viral phosphodiesterases for immune signaling evasion
Cyclic dinucleotides (CDNs) and other short oligonucleotides play fundamental roles in immune system activation in organisms ranging from bacteria to humans. In response, viruses use phosphodiesterase...
www.biorxiv.org
Sarah Gurev
@sarahgurev.bsky.social
· Aug 20
Sarah Gurev
@sarahgurev.bsky.social
· Aug 17

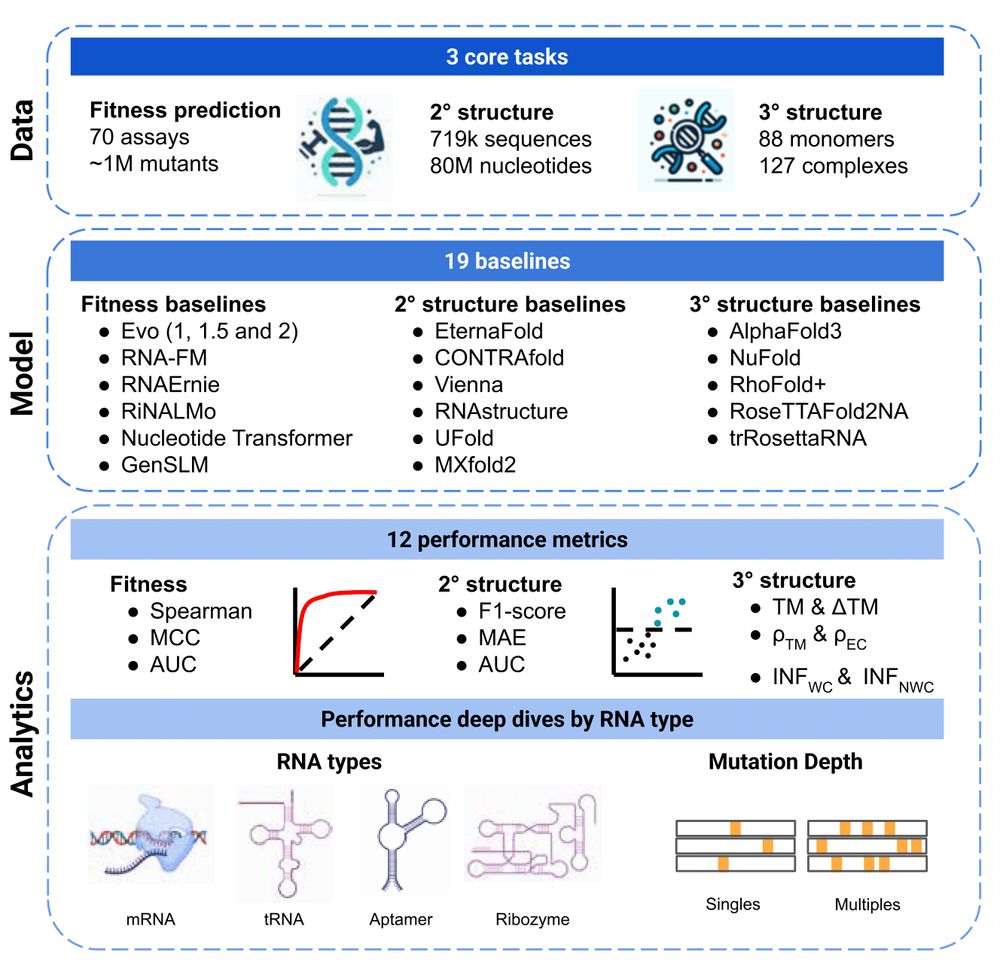
Variant effect prediction with reliability estimation across priority viruses
Viruses pose a significant threat to global health due to their rapid evolution, adaptability, and increasing potential for cross-species transmission. While advances in machine learning and the growi...
biorxiv.org
Sarah Gurev
@sarahgurev.bsky.social
· Aug 17
Sarah Gurev
@sarahgurev.bsky.social
· Aug 17
Sarah Gurev
@sarahgurev.bsky.social
· Aug 17
Sarah Gurev
@sarahgurev.bsky.social
· Aug 17
Sarah Gurev
@sarahgurev.bsky.social
· Aug 17
Reposted by Sarah Gurev
George Bouras
@gbouras13.bsky.social
· Aug 8

Protein Structure Informed Bacteriophage Genome Annotation with Phold
Bacteriophage (phage) genome annotation is essential for understanding their functional potential and suitability for use as therapeutic agents. Here we introduce Phold, an annotation framework utilis...
www.biorxiv.org
Reposted by Sarah Gurev
Yo Akiyama
@yoakiyama.bsky.social
· Aug 5

Scaling down protein language modeling with MSA Pairformer
Recent efforts in protein language modeling have focused on scaling single-sequence models and their training data, requiring vast compute resources that limit accessibility. Although models that use ...
biorxiv.org
Reposted by Sarah Gurev
Reposted by Sarah Gurev
Reposted by Sarah Gurev
Reposted by Sarah Gurev