Savage Lab
@savagecatsonly.bsky.social
320 followers
210 following
20 posts
Compartmentalizing your metabolism, and y’know, CRISPR stuff. Account managed by grad students and postdocs 🤙
@ucberkeleyofficial.bsky.social @innovativegenomics.bsky.social @hhmi.bsky.social
savagelab.org
Posts
Media
Videos
Starter Packs
Pinned
Reposted by Savage Lab
Doudna Lab
@doudna-lab.bsky.social
· Jul 29
Owen Tuck
@owentuck.bsky.social
· Jul 29

Recurrent acquisition of nuclease-protease pairs in antiviral immunity
Antiviral immune systems diversify by integrating new genes into existing pathways, creating new mechanisms of viral resistance. We identified genes encoding a predicted nuclease paired with a trypsin...
tinyurl.com
Reposted by Savage Lab
Reposted by Savage Lab
Ben Adler
@benadler.bsky.social
· Feb 26
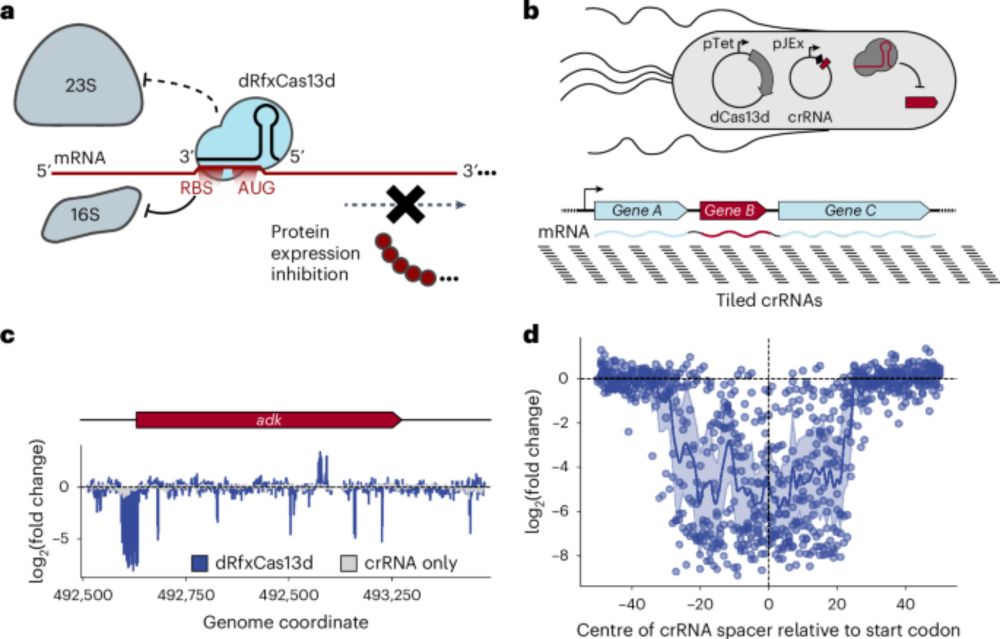
CRISPRi-ART enables functional genomics of diverse bacteriophages using RNA-binding dCas13d - Nature Microbiology
Leveraging RNA-targeting dCas13d enables selective interference with phage protein translation and facilitates measurement of phage gene fitness at a transcriptome-wide scale.
www.nature.com
Savage Lab
@savagecatsonly.bsky.social
· Feb 26

Latent activity in TnpB revealed by mutational scanning
TnpB is an evolutionarily diverse family of RNA-guided endonucleases associated with prokaryotic transposons. Due to their small size and putative evolutionary relationship to Cas12s, TnpB holds signi...
www.biorxiv.org
Reposted by Savage Lab
Savage Lab
@savagecatsonly.bsky.social
· Jan 24
Reposted by Savage Lab