Jaruwatana (Sodai) Lotharukpong
@sodail.bsky.social
140 followers
250 following
9 posts
ꙩ ꙫ ө ꚛ ꙮ ༗ :: complex multicellularity in brown algae :: doctoral researcher :: max planck institute for biology :: tübingen
https://lotharukpongjs.github.io/
Posts
Media
Videos
Starter Packs
Reposted by Jaruwatana (Sodai) Lotharukpong
Reposted by Jaruwatana (Sodai) Lotharukpong
Reposted by Jaruwatana (Sodai) Lotharukpong
Reposted by Jaruwatana (Sodai) Lotharukpong
Reposted by Jaruwatana (Sodai) Lotharukpong
Reposted by Jaruwatana (Sodai) Lotharukpong
Reposted by Jaruwatana (Sodai) Lotharukpong
Reposted by Jaruwatana (Sodai) Lotharukpong
Reposted by Jaruwatana (Sodai) Lotharukpong
Giulio Ermanno Pibiri
@jermp.bsky.social
· Aug 28

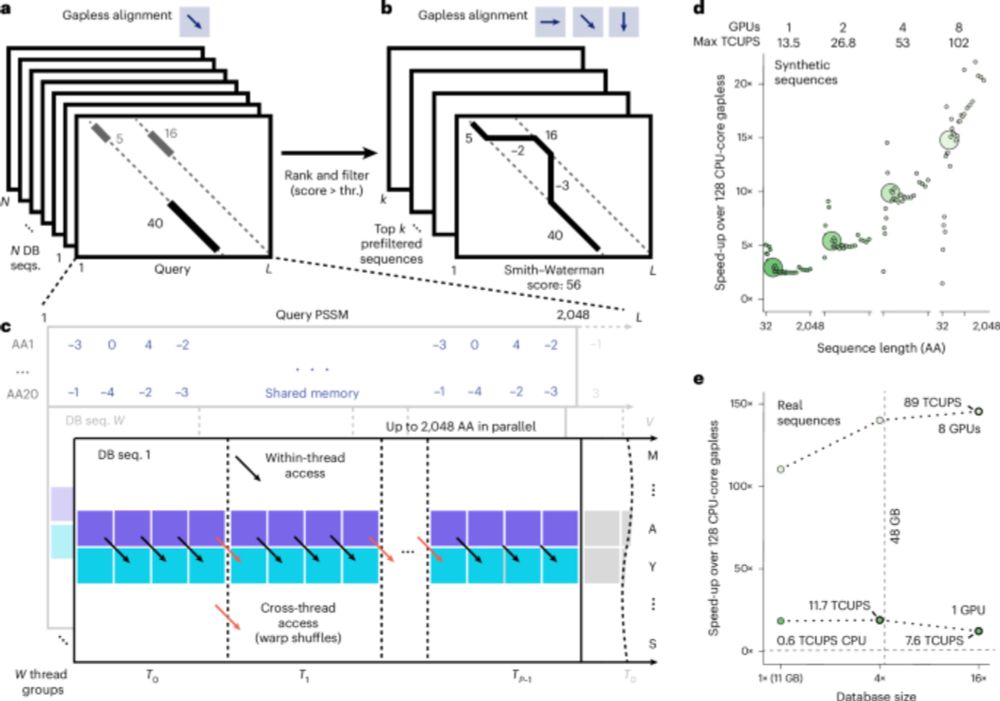
AllTheBacteria - all bacterial genomes assembled, available and searchable
The bacterial sequence data publicly available via the global DNA archives is a vast potential source of information on the evolution of bacteria. However, most of this sequence data is unassembled, o...
www.biorxiv.org
Reposted by Jaruwatana (Sodai) Lotharukpong
Yi-Jyun Luo
@yjluo.bsky.social
· Aug 18

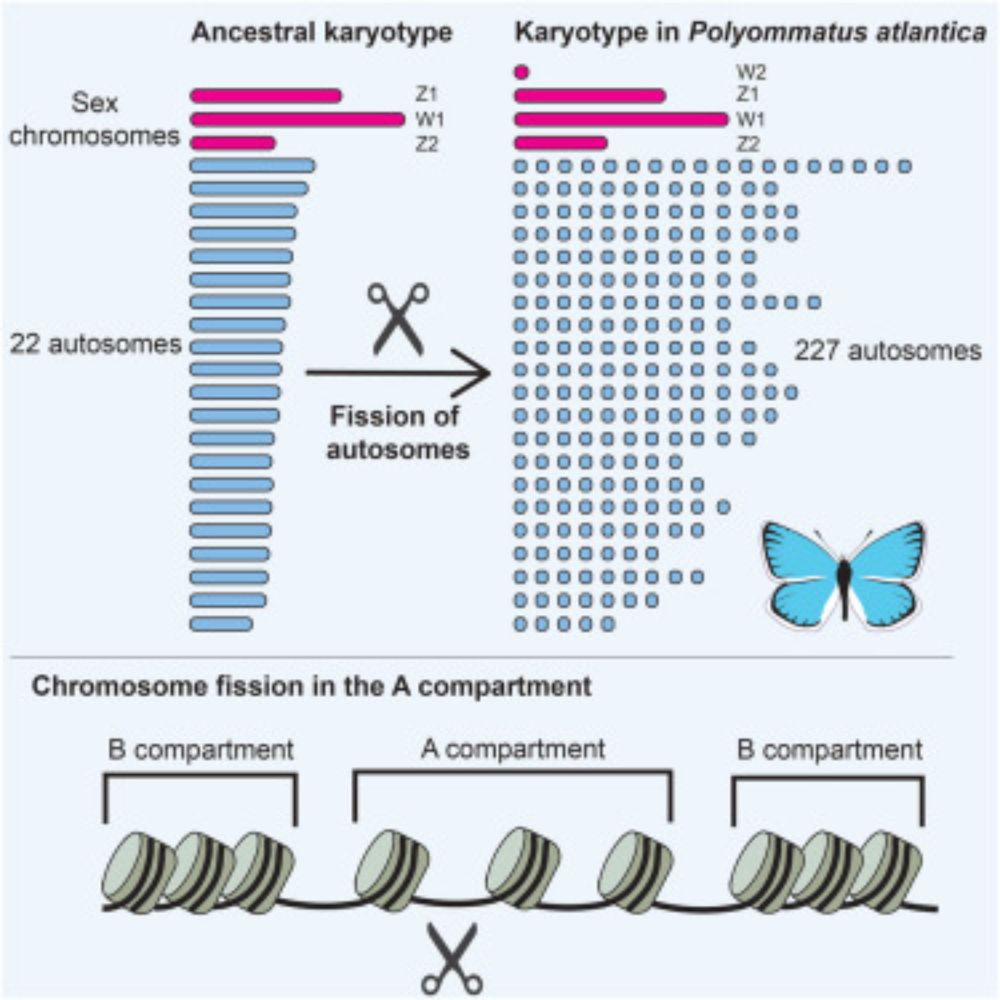
Conservation of bilaterian genome structure is the exception, not the rule - Genome Biology
Species from diverse animal lineages have conserved groups of orthologous genes together on the same chromosome for over half a billion years since the last common ancestor of bilaterians. Although no...
doi.org
Reposted by Jaruwatana (Sodai) Lotharukpong
Ferdinand Marlétaz
@ferdix.bsky.social
· Aug 13

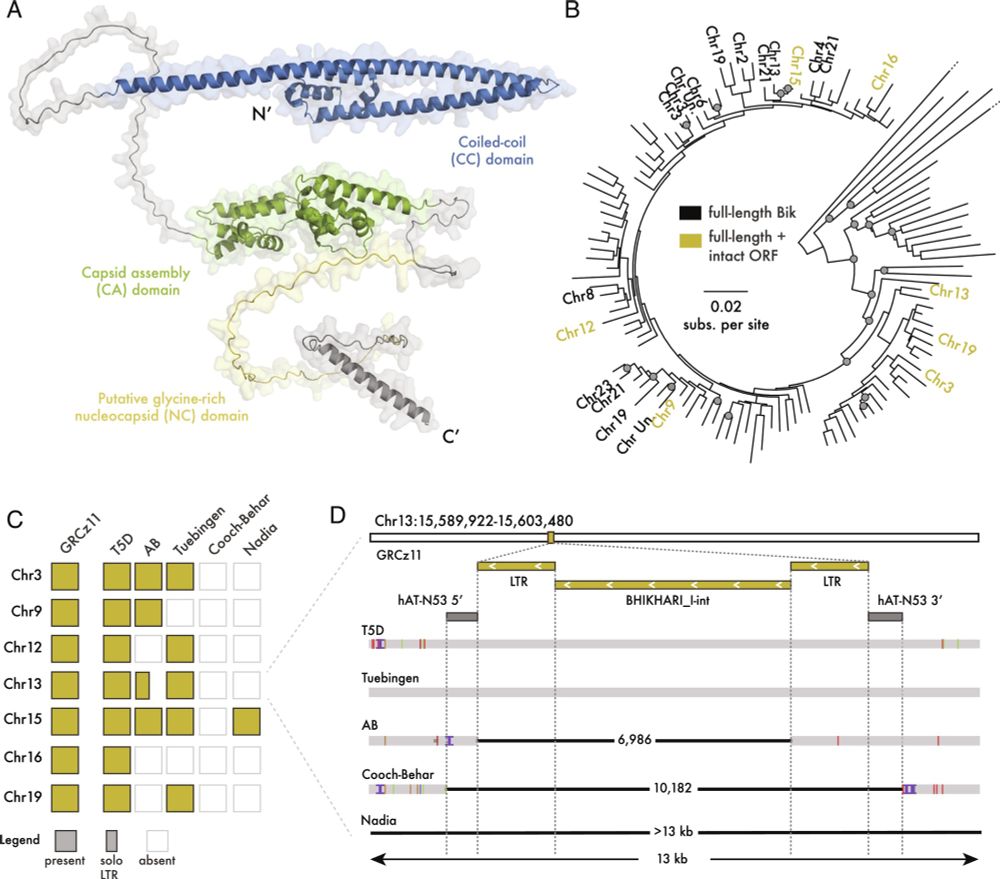
The genomic origin of the unique chaetognath body plan - Nature
Genomic, single-cell transcriptomic and epigenetic analyses show that chaetognaths, following extensive gene loss in the gnathiferan lineage, relied on newly evolved genes and lineage-specific tandem ...
www.nature.com
Reposted by Jaruwatana (Sodai) Lotharukpong
Reposted by Jaruwatana (Sodai) Lotharukpong
Iana V. Kim
@ianakim.bsky.social
· May 7
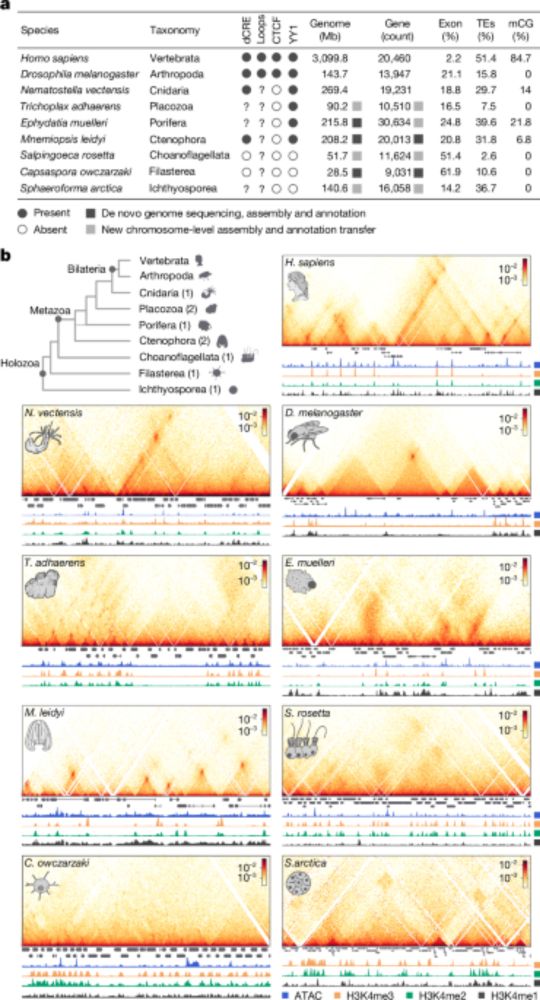
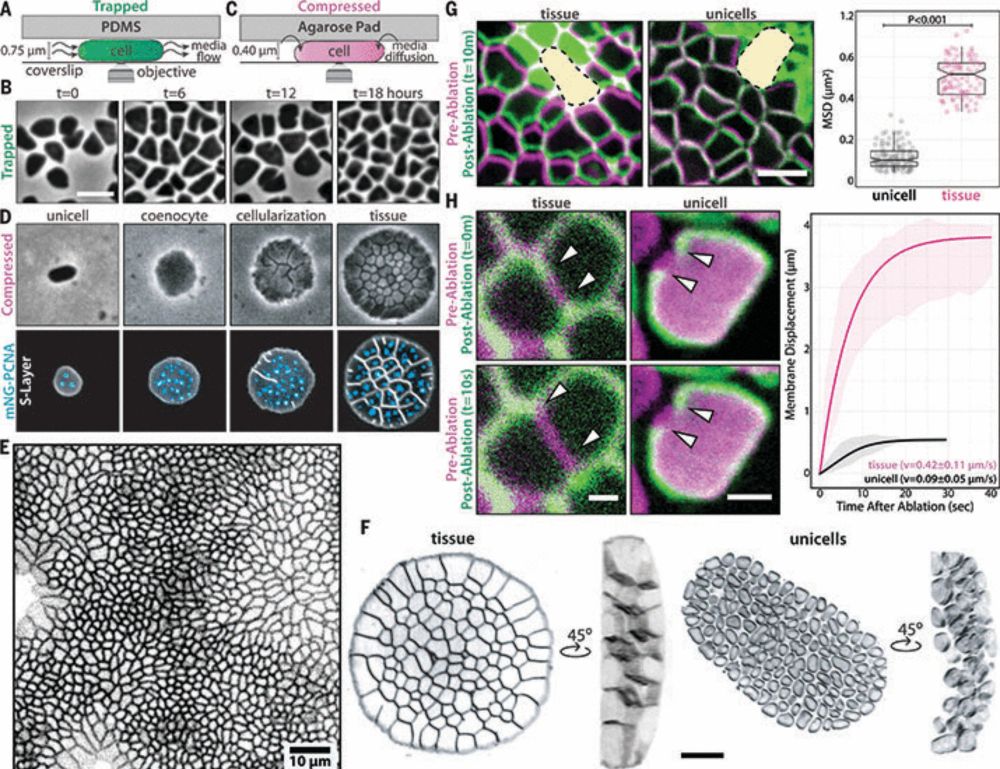
Chromatin loops are an ancestral hallmark of the animal regulatory genome - Nature
The physical organization of the genome in non-bilaterian animals and their closest unicellular relatives is characterized; comparative analysis shows chromatin looping is a conserved feature of ...
www.nature.com
Reposted by Jaruwatana (Sodai) Lotharukpong
Heng Li
@lh3lh3.bsky.social
· Apr 18

Efficient near telomere-to-telomere assembly of Nanopore Simplex reads
Telomere-to-telomere (T2T) assembly is the ultimate goal for de novo genome assembly. Existing algorithms capable of near T2T assembly all require Oxford Nanopore Technologies (ONT) ultra-long reads w...
www.biorxiv.org
Reposted by Jaruwatana (Sodai) Lotharukpong
Reposted by Jaruwatana (Sodai) Lotharukpong
Reposted by Jaruwatana (Sodai) Lotharukpong
Reposted by Jaruwatana (Sodai) Lotharukpong