Deni Szokoli
@szokoli.bsky.social
34 followers
53 following
27 posts
Studying ancient ribozymes. Likes group II introns a little too much. PhD @mutschlerlab.bsky.social
Posts
Media
Videos
Starter Packs
Pinned
Deni Szokoli
@szokoli.bsky.social
· Jul 2
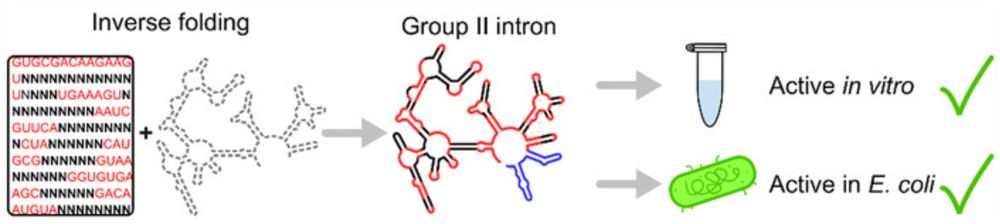
Computational De Novo Design of Group II Introns Yields Highly Active Ribozymes
Group II Introns (G2Is) are large self-splicing ribozymes with promising biotechnological applications. This study utilized RNA inverse folding to design three novel G2Is. The designed intron Arq.I2,...
doi.org
Reposted by Deni Szokoli
Reposted by Deni Szokoli
Reposted by Deni Szokoli
Deni Szokoli
@szokoli.bsky.social
· Jul 3
Deni Szokoli
@szokoli.bsky.social
· Jul 3
Deni Szokoli
@szokoli.bsky.social
· Jul 2
Deni Szokoli
@szokoli.bsky.social
· Jul 2
Deni Szokoli
@szokoli.bsky.social
· Jul 2
Deni Szokoli
@szokoli.bsky.social
· Jul 2
Deni Szokoli
@szokoli.bsky.social
· Jul 2
Deni Szokoli
@szokoli.bsky.social
· Jul 2
Deni Szokoli
@szokoli.bsky.social
· Jul 2

Computational De Novo Design of Group II Introns Yields Highly Active Ribozymes
Group II Introns (G2Is) are large self-splicing ribozymes with promising biotechnological applications. This study utilized RNA inverse folding to design three novel G2Is. The designed intron Arq.I2,...
doi.org
Reposted by Deni Szokoli
Reposted by Deni Szokoli
Phil Holliger
@philholliger.bsky.social
· May 29
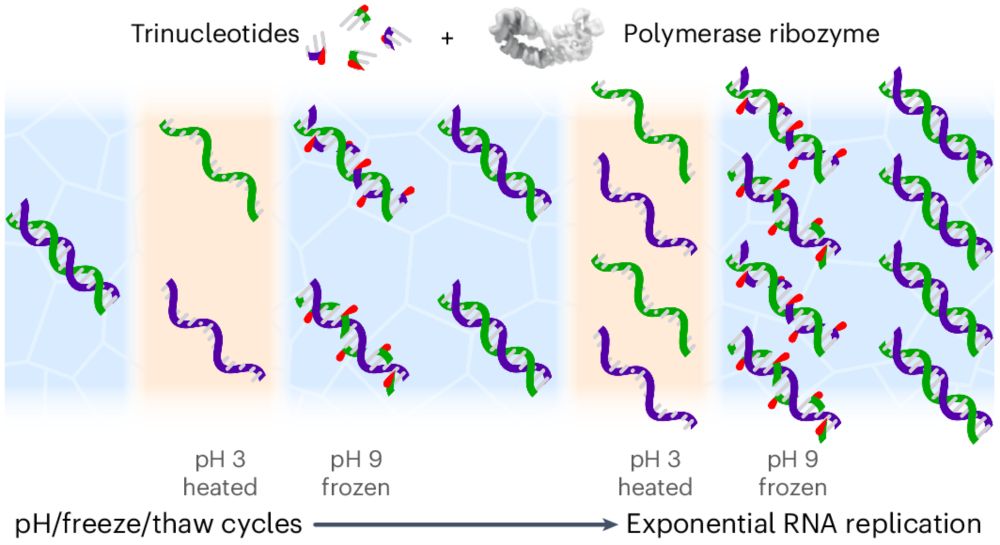
Trinucleotide substrates under pH–freeze–thaw cycles enable open-ended exponential RNA replication by a polymerase ribozyme
Nature Chemistry - Models of abiotic RNA replication suffer from inherent product inhibition arising from the high stability of RNA duplexes. Now, it has been shown that RNA trinucleotide...
rdcu.be
Deni Szokoli
@szokoli.bsky.social
· May 26

An intron endonuclease facilitates interference competition between coinfecting viruses
Introns containing homing endonucleases are widespread in nature and have long been assumed to be selfish elements that provide no benefit to the host organism. These genetic elements are common in vi...
www.science.org
Deni Szokoli
@szokoli.bsky.social
· May 26
Liana Merk
@bacteriyay.bsky.social
· May 24

Prevalence of Group II Introns in Phage Genomes
Although bacteriophage genomes are under strong selective pressure for high coding density, they are still frequently invaded by mobile genetic elements (MGEs). Group II introns are MGEs that reduce h...
www.biorxiv.org
Reposted by Deni Szokoli








