Tara Bartolec
@tarabartolec.bsky.social
140 followers
160 following
11 posts
EIPOD-LinC postdoc fellow in the Savitski and Typas labs at EMBL, Heidelberg. Proteomics, phages, PTMs, XLMS 🇦🇺🦠🧑🔬
Posts
Media
Videos
Starter Packs
Pinned
Reposted by Tara Bartolec
Ruoshi
@ruoshiz.bsky.social
· 22d
Reposted by Tara Bartolec
Reposted by Tara Bartolec
Savitski Lab
@savitski-lab.bsky.social
· Apr 29

High-throughput peptide-centric local stability assay extends protein-ligand identification to membrane proteins, tissues, and bacteria
Systematic mapping of protein-ligand interactions is essential for understanding biological processes and drug mechanisms. Peptide-centric local stability assay (PELSA) is a powerful tool for detectin...
www.biorxiv.org
Reposted by Tara Bartolec
Reposted by Tara Bartolec
Reposted by Tara Bartolec
Reposted by Tara Bartolec
Reposted by Tara Bartolec
Ben Adler
@benadler.bsky.social
· Feb 26
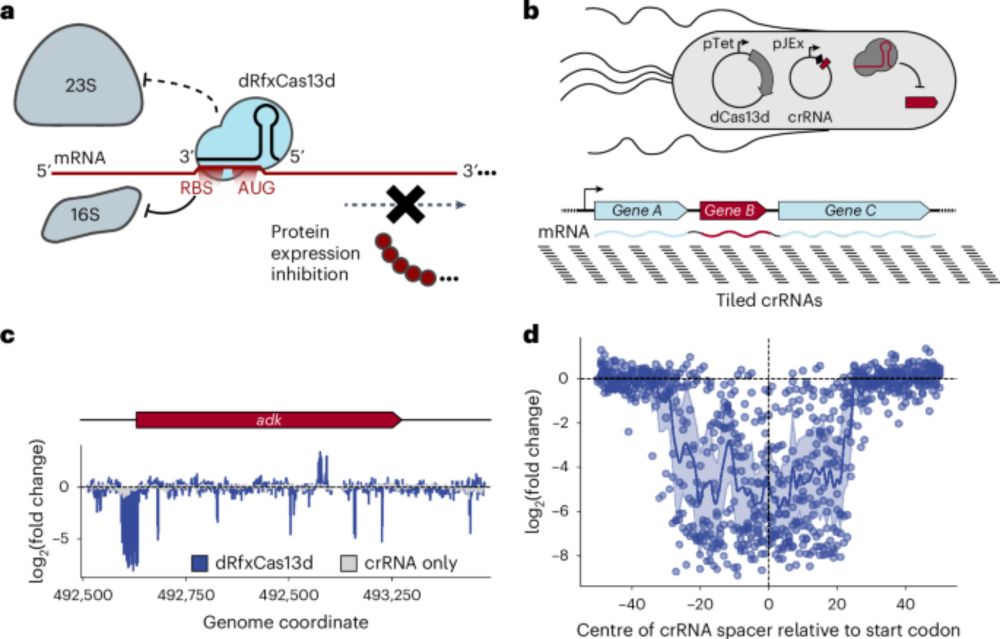
CRISPRi-ART enables functional genomics of diverse bacteriophages using RNA-binding dCas13d - Nature Microbiology
Leveraging RNA-targeting dCas13d enables selective interference with phage protein translation and facilitates measurement of phage gene fitness at a transcriptome-wide scale.
www.nature.com
Reposted by Tara Bartolec
Reposted by Tara Bartolec
Aude Bernheim
@audeber.bsky.social
· Feb 19
Webservice | DefenseFinder webservice and knowledge base
On this site, you can freely use (without any login) the DefenseFinder webservice (see below) and get help to navigate the ever expanding world of defense systems.There is a collaborative knowledge ba...
defensefinder.mdmlab.fr
Reposted by Tara Bartolec
Reposted by Tara Bartolec
Reposted by Tara Bartolec
Reposted by Tara Bartolec
Typas Lab
@typaslab.bsky.social
· Dec 23

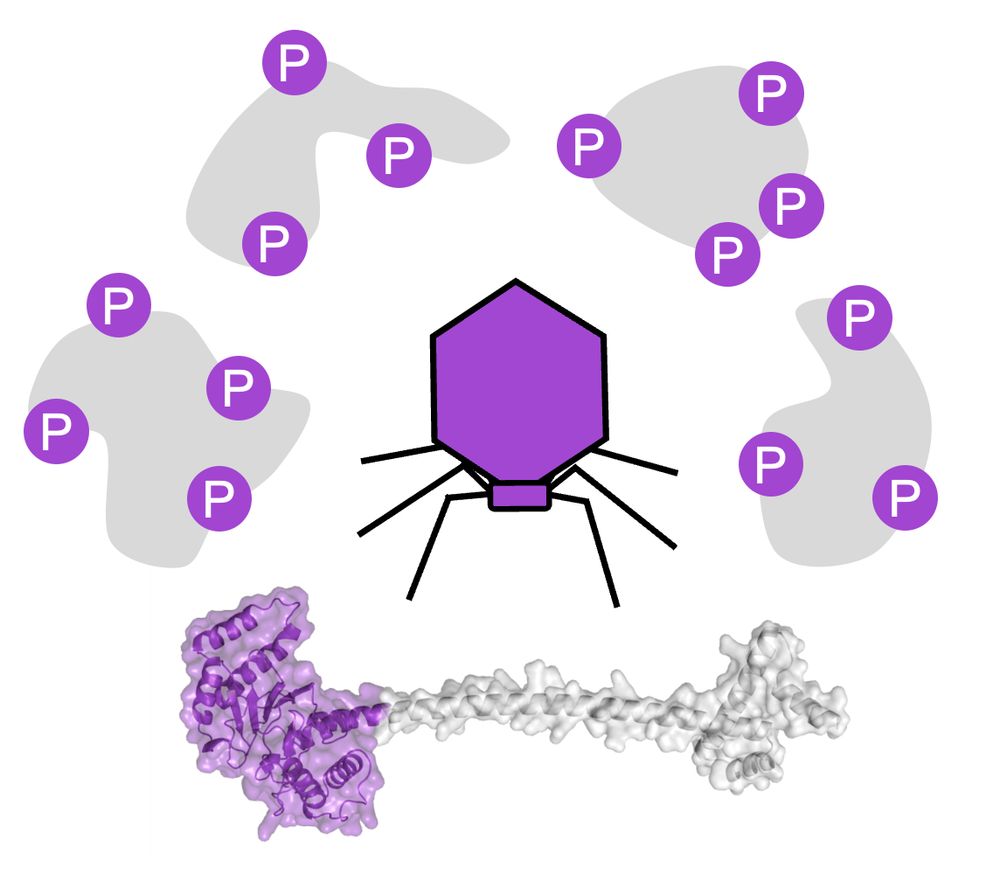
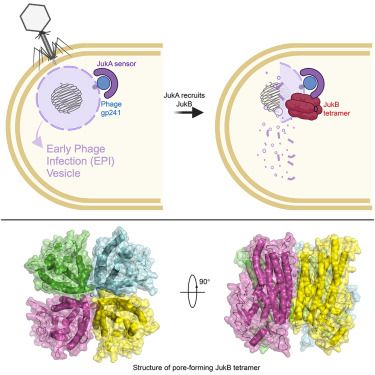
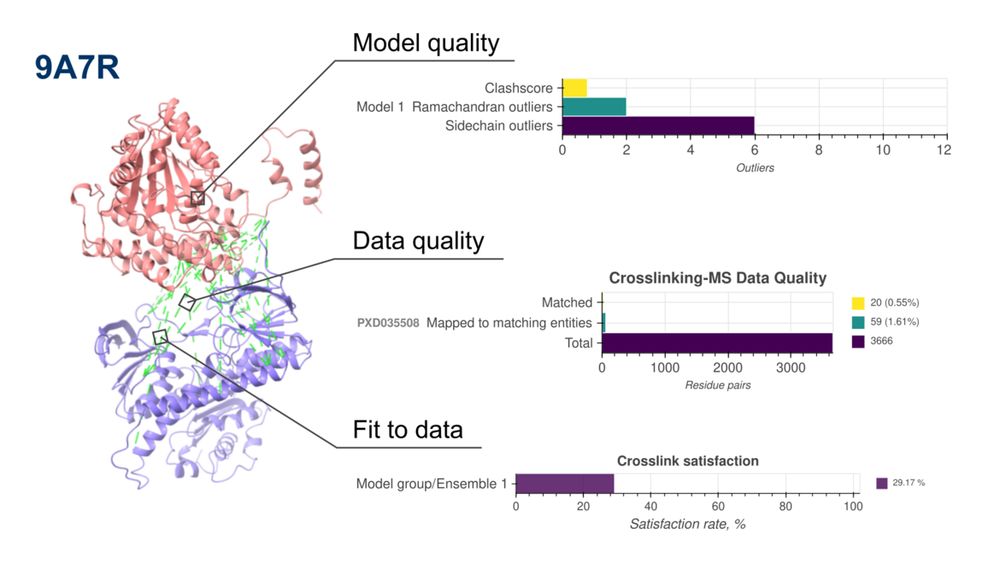
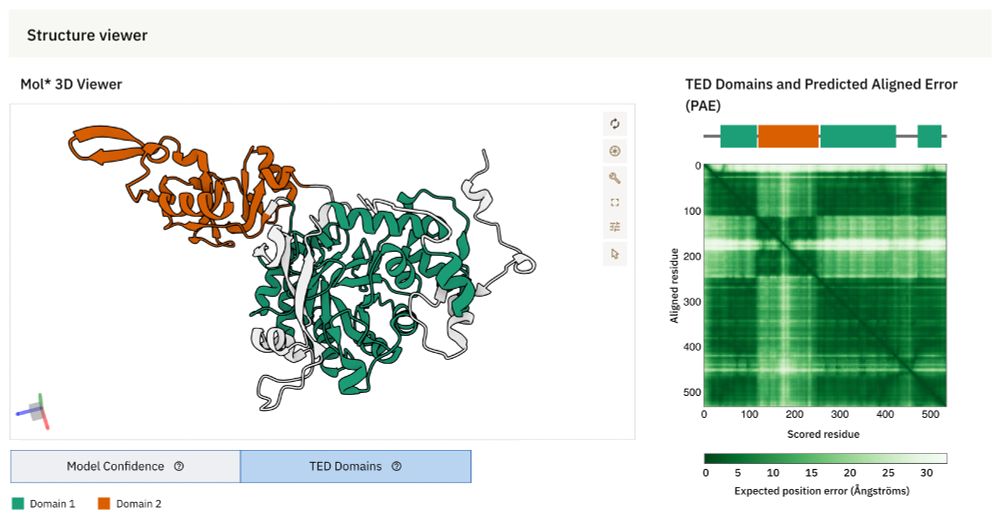
Pervasive phosphorylation by phage T7 kinase disarms bacterial defenses
Bacteria and bacteriophages are in a constant arms race to develop bacterial defense and phage counter-defense systems. Currently known phage counter-defense systems are specific to (the activity of) ...
www.biorxiv.org
Tara Bartolec
@tarabartolec.bsky.social
· Dec 23

Pervasive phosphorylation by phage T7 kinase disarms bacterial defenses
Bacteria and bacteriophages are in a constant arms race to develop bacterial defense and phage counter-defense systems. Currently known phage counter-defense systems are specific to (the activity of) ...
www.biorxiv.org
Tara Bartolec
@tarabartolec.bsky.social
· Dec 23
Tara Bartolec
@tarabartolec.bsky.social
· Dec 23
Tara Bartolec
@tarabartolec.bsky.social
· Dec 23
Tara Bartolec
@tarabartolec.bsky.social
· Dec 23
Tara Bartolec
@tarabartolec.bsky.social
· Dec 23
Tara Bartolec
@tarabartolec.bsky.social
· Dec 23
Tara Bartolec
@tarabartolec.bsky.social
· Dec 23