Tom Dongmin Kim
@tomdongmin.bsky.social
170 followers
720 following
2 posts
Postdoc @Cardonalab, protein engineering, directed evolution, photosystem II, photosynthesis, HTS, genomics, structural bioinformatics, CRISPR, machine learning
Posts
Media
Videos
Starter Packs
Reposted by Tom Dongmin Kim
Tanai Cardona
@tanaicardona.bsky.social
· May 28

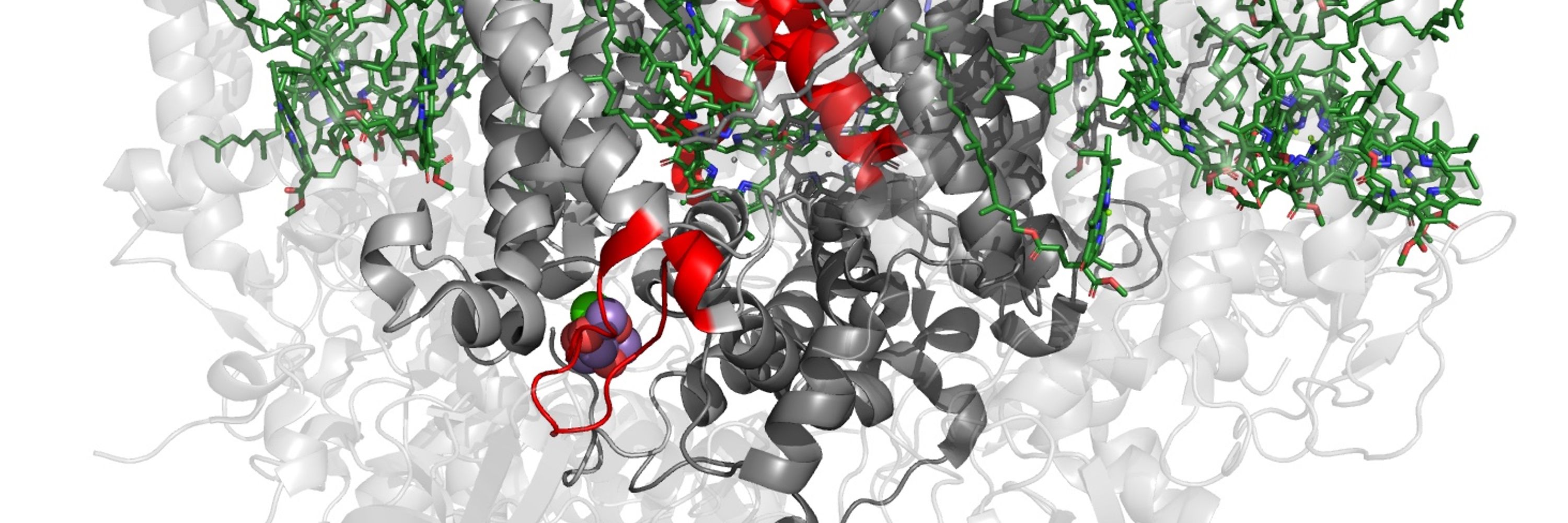
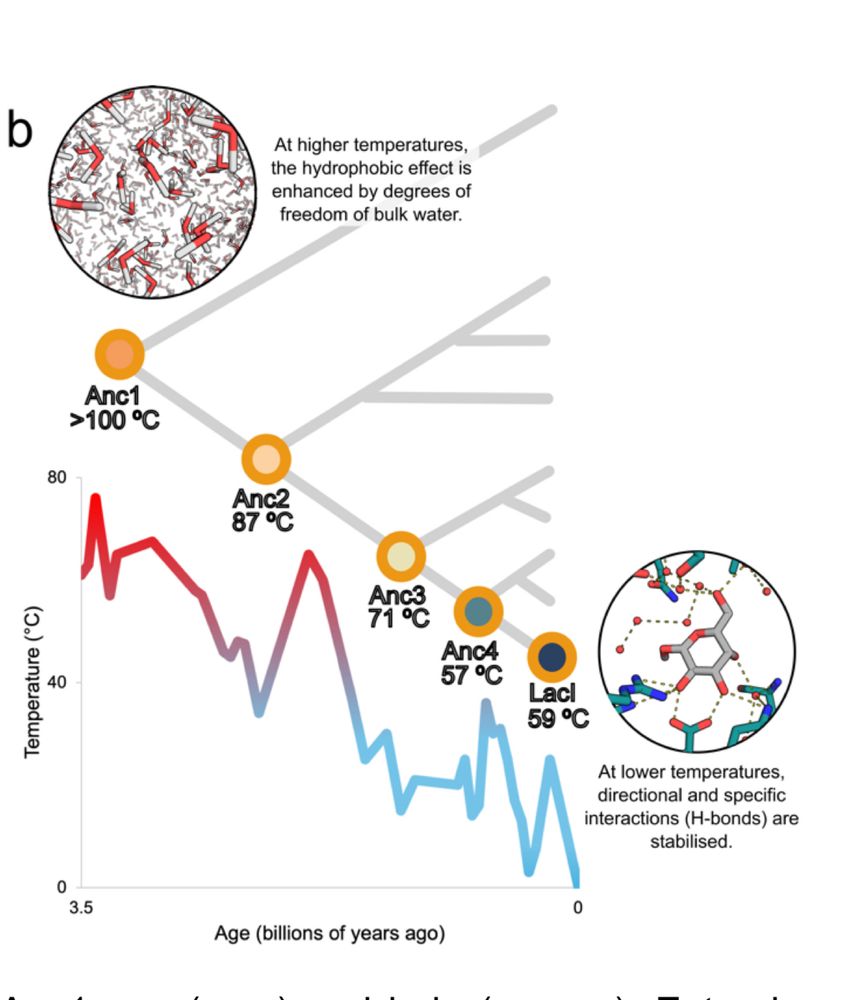
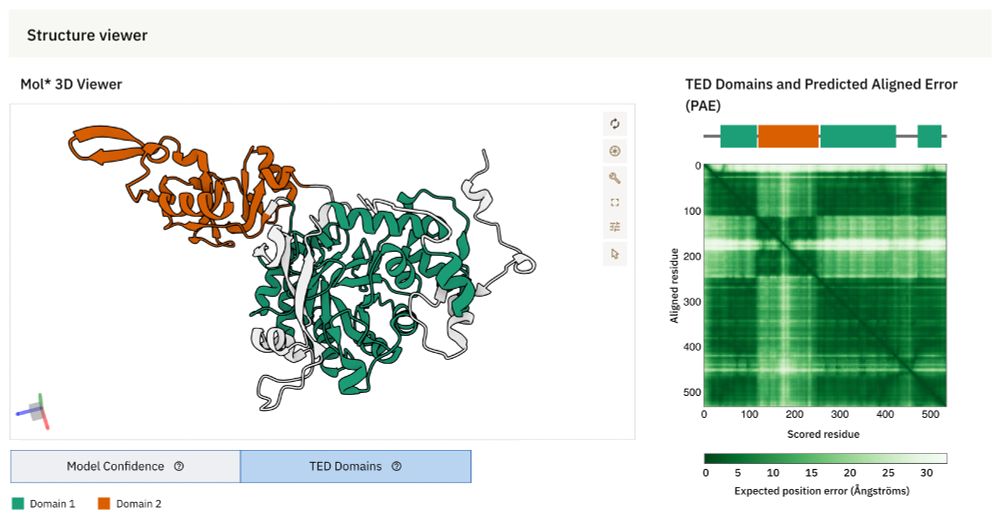
Exploring the Structural Diversity and Evolution of the D1 Subunit of Photosystem II Using AlphaFold and Foldtree
Although our knowledge of photosystem II has expanded to include time-resolved atomic details, the diversity of experimental structures of the enzyme remains limited. Recent advances in protein struc...
onlinelibrary.wiley.com
Reposted by Tom Dongmin Kim
Reposted by Tom Dongmin Kim
Vega Shah
@dr-alphalyrae.bsky.social
· Apr 6

Siemens Agrees to Buy Dotmatics for $5.1 Billion, Expanding AI Portfolio to Life Sciences
The deal extends Siemens’s artificial-intelligence-powered product-lifecycle-management portfolio and increases the company’s total addressable market in industrial software by $11 billion.
www.wsj.com
Reposted by Tom Dongmin Kim
Reposted by Tom Dongmin Kim
Reposted by Tom Dongmin Kim
Reposted by Tom Dongmin Kim
Reposted by Tom Dongmin Kim
Reposted by Tom Dongmin Kim
Reposted by Tom Dongmin Kim
Reposted by Tom Dongmin Kim
Cyanotrans
@cyanotrans.bsky.social
· Mar 28

Enhancing the production of chlorophyll f in the cyanobacterium Synechocystis sp. PCC 6803
One potential approach to improve the productivity of cyanobacteria and microalgae is to enhance photosynthetic efficiency by introducing far-red absorbing pigment molecules (such as chlorophylls f a...
onlinelibrary.wiley.com
Reposted by Tom Dongmin Kim
Reposted by Tom Dongmin Kim
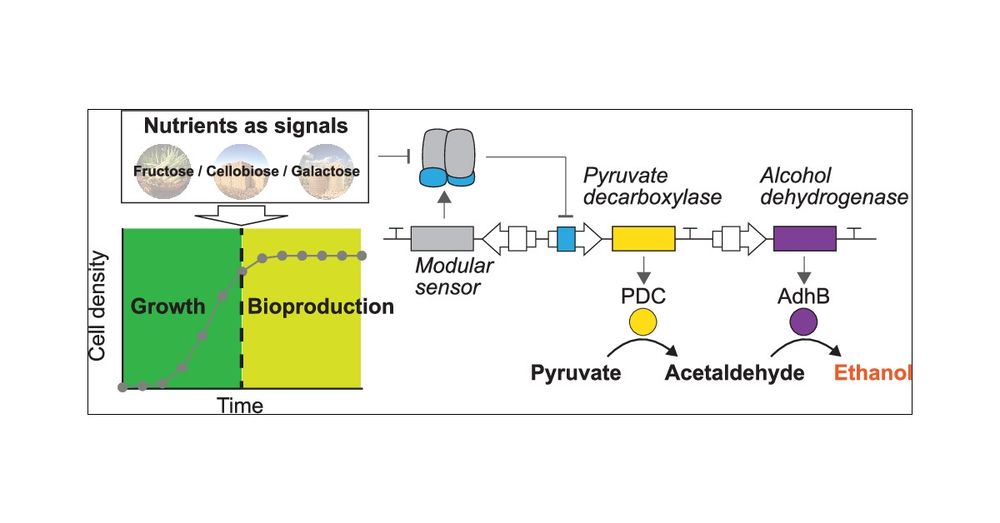
Olivier Borkowski
@oborkowski.bsky.social
· Mar 29
Reposted by Tom Dongmin Kim
Reposted by Tom Dongmin Kim
Reposted by Tom Dongmin Kim
Reposted by Tom Dongmin Kim
Reposted by Tom Dongmin Kim
Bird account
@dialecticbio.bsky.social
· Dec 28
GitHub - Graylab/DL4Proteins-notebooks: Colab Notebooks covering deep learning tools for biomolecular structure prediction and design
Colab Notebooks covering deep learning tools for biomolecular structure prediction and design - Graylab/DL4Proteins-notebooks
github.com
Reposted by Tom Dongmin Kim
Reposted by Tom Dongmin Kim
Loren Williams
@lorendw.bsky.social
· Mar 8
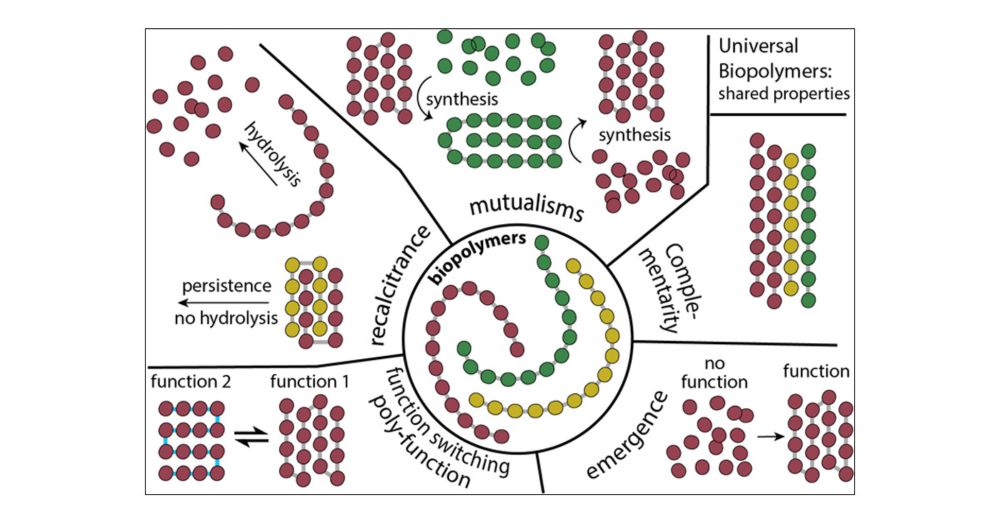
Biological Polymers: Evolution, Function, and Significance
ConspectusA holistic description of biopolymers and their evolutionary origins will contribute to our understanding of biochemistry, biology, the origins of life, and signatures of life outside our pl...
pubs.acs.org