Toufic Mayassi
@touficmayassi.bsky.social
280 followers
150 following
23 posts
Mucosal immunologist drawn to the computational world | Bana Jabri lab graduate | Postdoc in @thexavierlab.bsky.social at the @broadinstitute.bsky.social
Posts
Media
Videos
Starter Packs
Pinned
Toufic Mayassi
@touficmayassi.bsky.social
· Nov 21
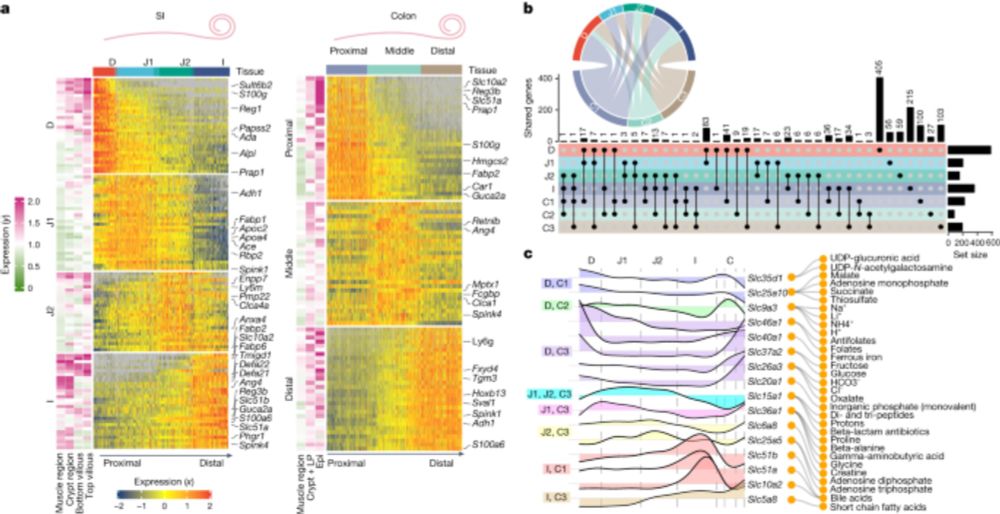
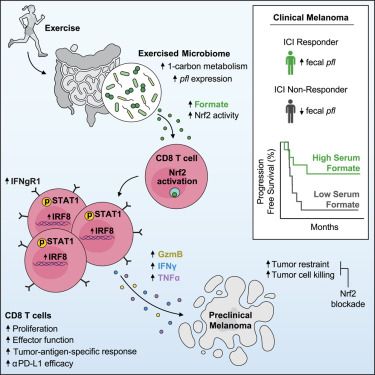
Spatially restricted immune and microbiota-driven adaptation of the gut - Nature
Intestinal regionalization is characterized by robust and resilient structural cell states and the intestine can adapt to environmental stress in a spatially controlled manner through crosstalk betwee...
www.nature.com
Reposted by Toufic Mayassi
Toufic Mayassi
@touficmayassi.bsky.social
· Nov 24
Reposted by Toufic Mayassi
Ivan Zanoni
@lozanzi.bsky.social
· Nov 23
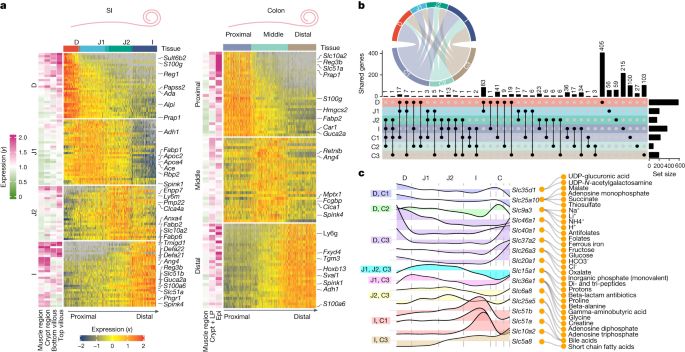
Spatially restricted immune and microbiota-driven adaptation of the gut - Nature
Intestinal regionalization is characterized by robust and resilient structural cell states and the intestine can adapt to environmental stress in a spatially controlled manner through crosstalk betwee...
www.nature.com
Toufic Mayassi
@touficmayassi.bsky.social
· Nov 21
Toufic Mayassi
@touficmayassi.bsky.social
· Nov 21
Toufic Mayassi
@touficmayassi.bsky.social
· Nov 21
Toufic Mayassi
@touficmayassi.bsky.social
· Nov 21
Toufic Mayassi
@touficmayassi.bsky.social
· Nov 21
Toufic Mayassi
@touficmayassi.bsky.social
· Nov 21