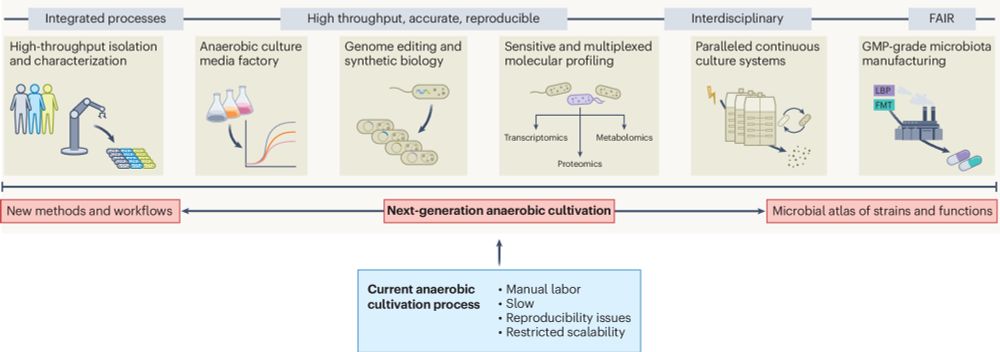
www.biorxiv.org/content/10.1...
www.biorxiv.org/content/10.1...
Congratulations to Dina Kao @ualberta.bsky.social & team!
Gift link:
authors.elsevier.com/c/1lphg8nByr...
Congratulations to Dina Kao @ualberta.bsky.social & team!
Gift link:
authors.elsevier.com/c/1lphg8nByr...
Thanks for spreading the word.
#phdchat #phdlife #academiclife
1️⃣ Microbiome signatures of early on-set colorectal cancer
2️⃣ High-resolution computational metagenomics for the study of human microbiome transmission
More info: www.semm.it/education/op...
Apply by June 9!
Thanks for spreading the word.
#phdchat #phdlife #academiclife
Commentary in @cellcellpress.bsky.social!
by @msabrysarhan.bsky.social Giacomo Antonello @whansi.bsky.social @cengoni.bsky.social @dmascalzoni.bsky.social @leviwaldron1.bsky.social Nicola Segata, Christian Fuchsberger
www.cell.com/cell/fulltex...
1/4
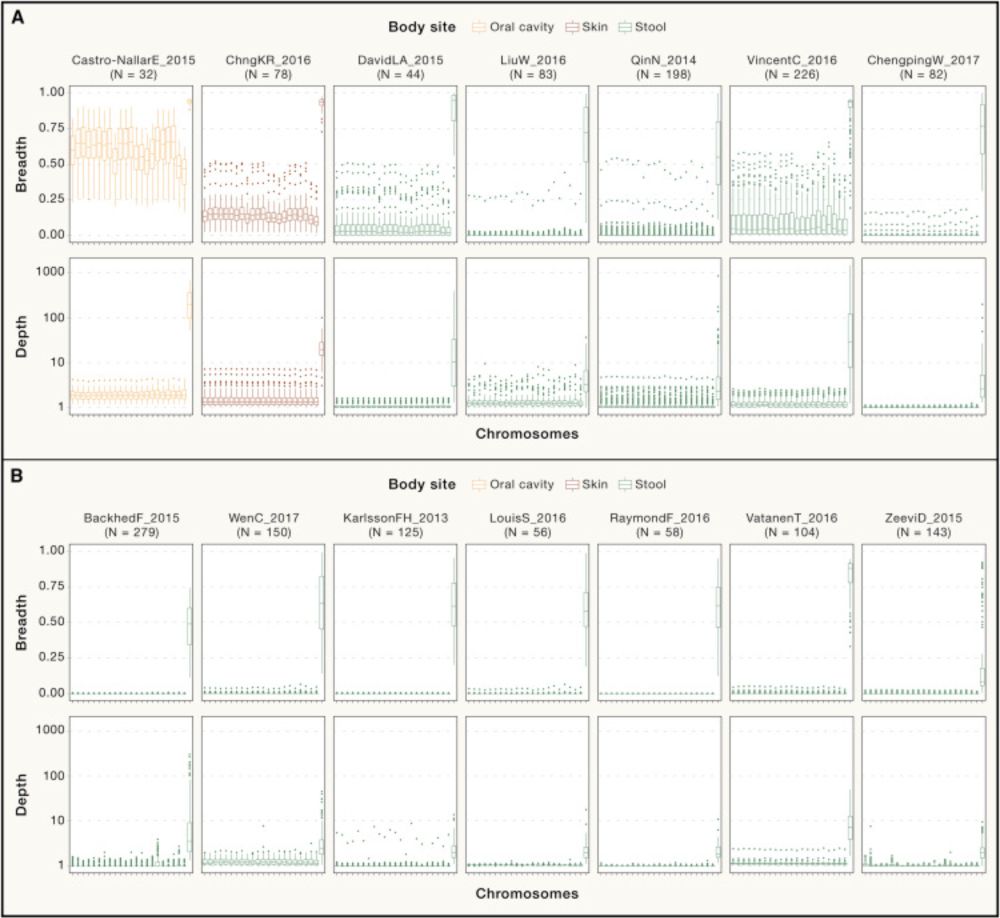
Commentary in @cellcellpress.bsky.social!
by @msabrysarhan.bsky.social Giacomo Antonello @whansi.bsky.social @cengoni.bsky.social @dmascalzoni.bsky.social @leviwaldron1.bsky.social Nicola Segata, Christian Fuchsberger
www.cell.com/cell/fulltex...
1/4
Fascinating translational science in action!
@brunslab.bsky.social ky.social @easlnews.bsky.social ky.social
#scRNAseq #ACLF
Fascinating translational science in action!
@brunslab.bsky.social ky.social @easlnews.bsky.social ky.social
#scRNAseq #ACLF
📅 16 – 19 Sep 2025
📍 EMBL Heidelberg and Virtual
📥 Submit your abstract by 24 June
➡️ https://s.embl.org/ees25-08-bl

📅 16 – 19 Sep 2025
📍 EMBL Heidelberg and Virtual
📥 Submit your abstract by 24 June
➡️ https://s.embl.org/ees25-08-bl