Tom Williams
@tweethinking.bsky.social
150 followers
79 following
14 posts
Computational evolutionary biologist - phylogenetics, molecular and microbial evolution
Posts
Media
Videos
Starter Packs
Reposted by Tom Williams
Reposted by Tom Williams
Sishuo Wang
@sishuowang.bsky.social
· Jul 11
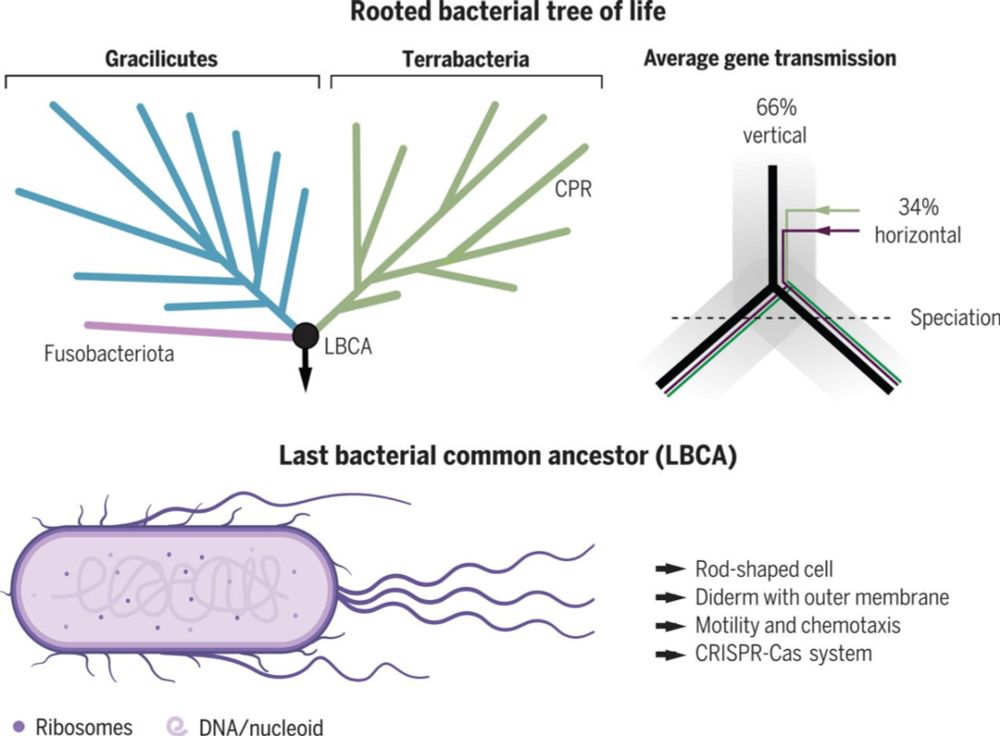
New Ways to Root Phylogenomic trees: the Smart, the symbiotic, the violent
To root a phylogenomic tree is very important to study the evolutionary relationship between species. However, typically the substitution models (say GTR, LG, etc.) people use are time-reversible m…
sishuowang2022.wordpress.com
Tom Williams
@tweethinking.bsky.social
· Jun 17
DEEMteam_Orsay
@deemteam.bsky.social
· Jun 17
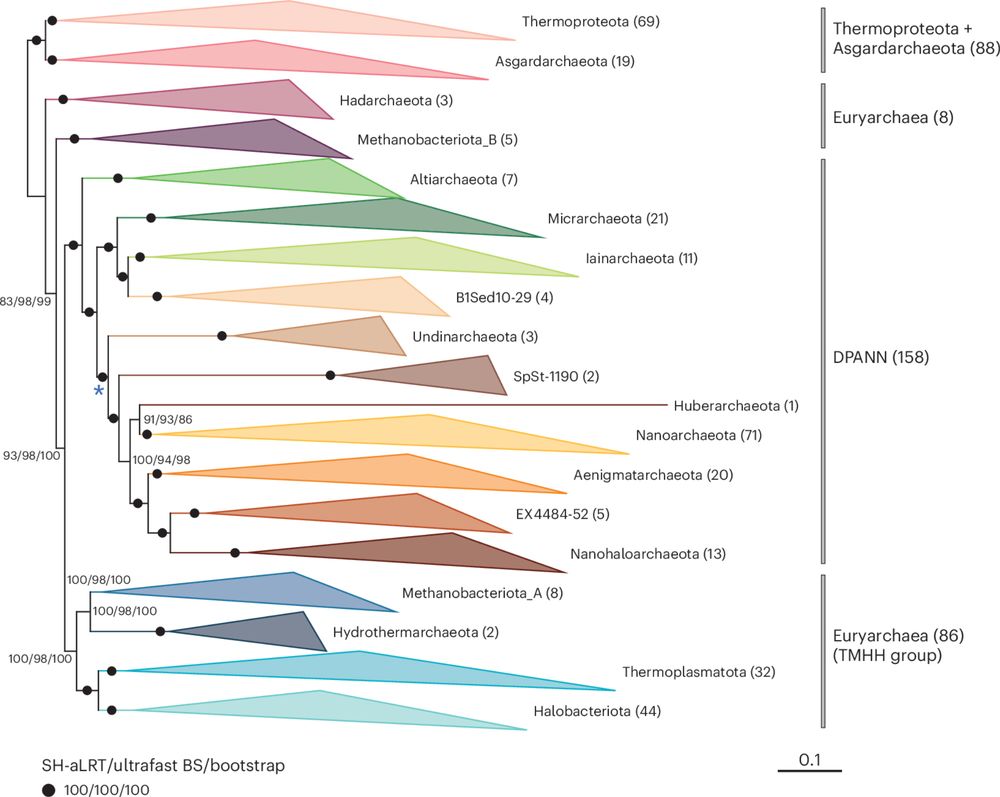
Phylogenomic analyses indicate the archaeal superphylum DPANN originated from free-living euryarchaeal-like ancestors
Nature Microbiology - Phylogenetic reconstructions with conserved protein markers from the 11 known DPANN phyla reveal their monophyletic placement within the Euryarchaeota.
rdcu.be
Reposted by Tom Williams
Reposted by Tom Williams
Reposted by Tom Williams
Alexis Stamatakis
@stamatak.bsky.social
· Mar 19

raxtax: A k-mer-based non-Bayesian Taxonomic Classifier
Taxonomic classification in biodiversity studies is the process of assigning the anonymous sequences of a marker gene (barcode) to a specific lineage using a reference database that contains named seq...
www.biorxiv.org
Tom Williams
@tweethinking.bsky.social
· Mar 19
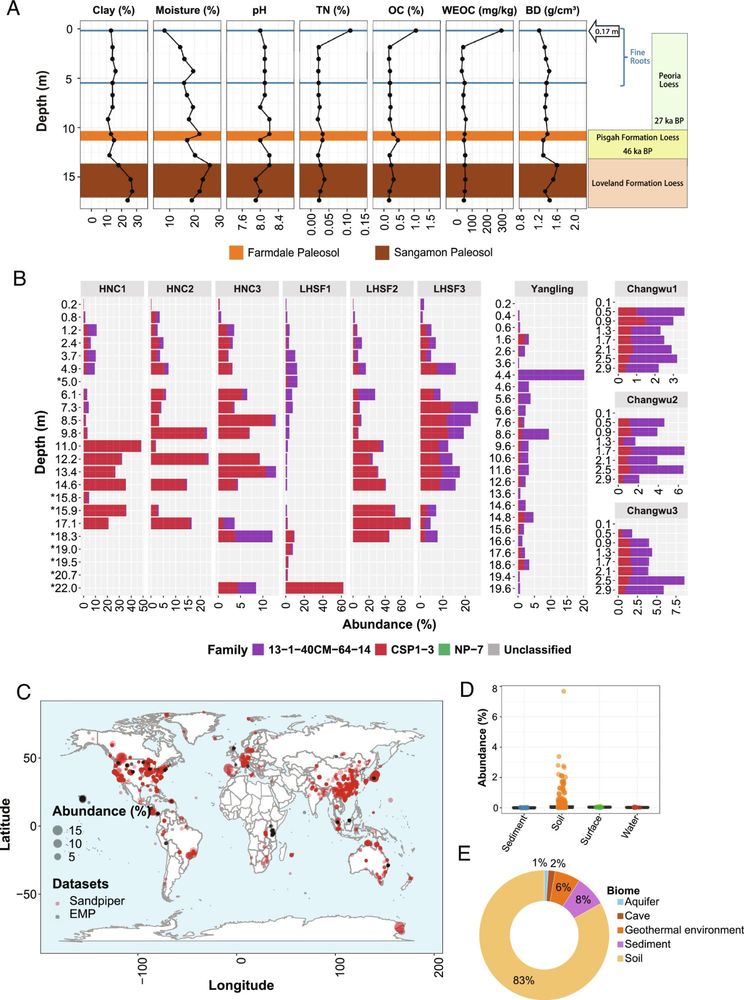
Diversification, niche adaptation, and evolution of a candidate phylum thriving in the deep Critical Zone | PNAS
The deep subsurface soil microbiome encompasses a vast amount of understudied phylogenetic
diversity and metabolic novelty, and the metabolic capab...
www.pnas.org
Tom Williams
@tweethinking.bsky.social
· Mar 12
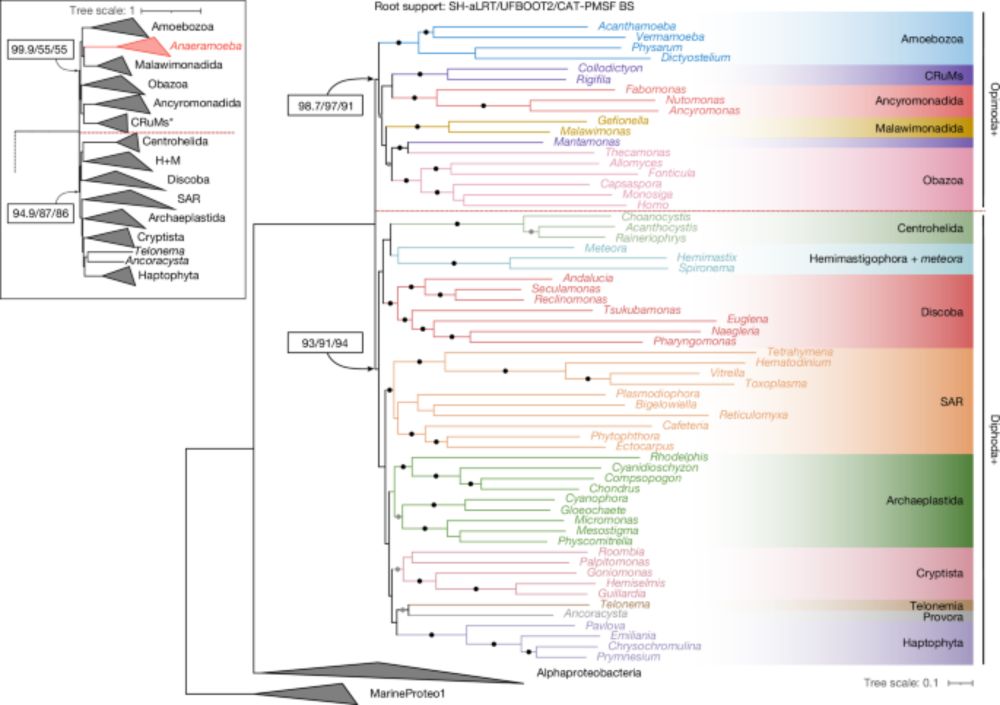
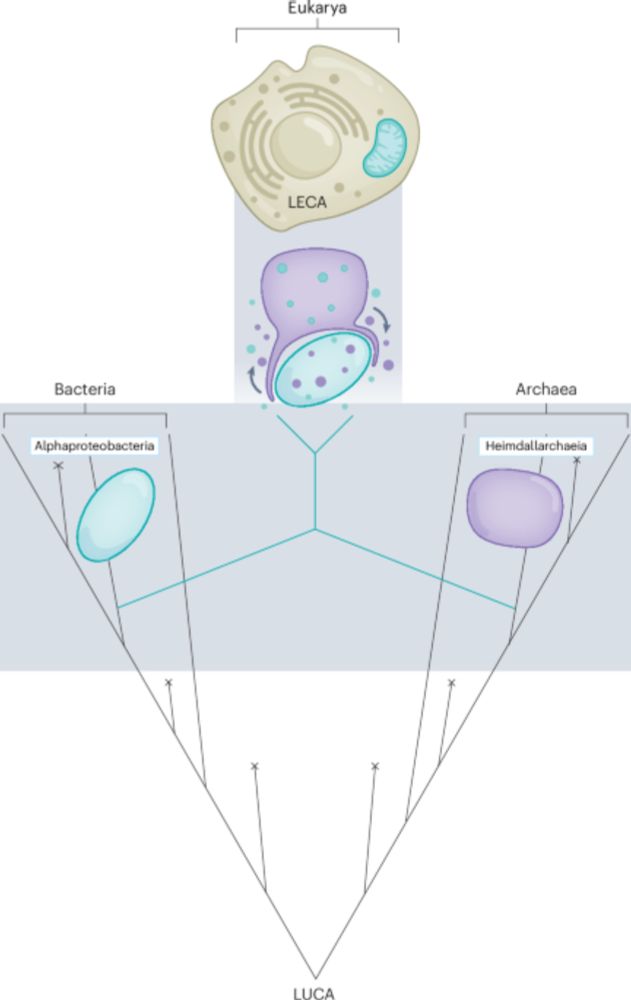
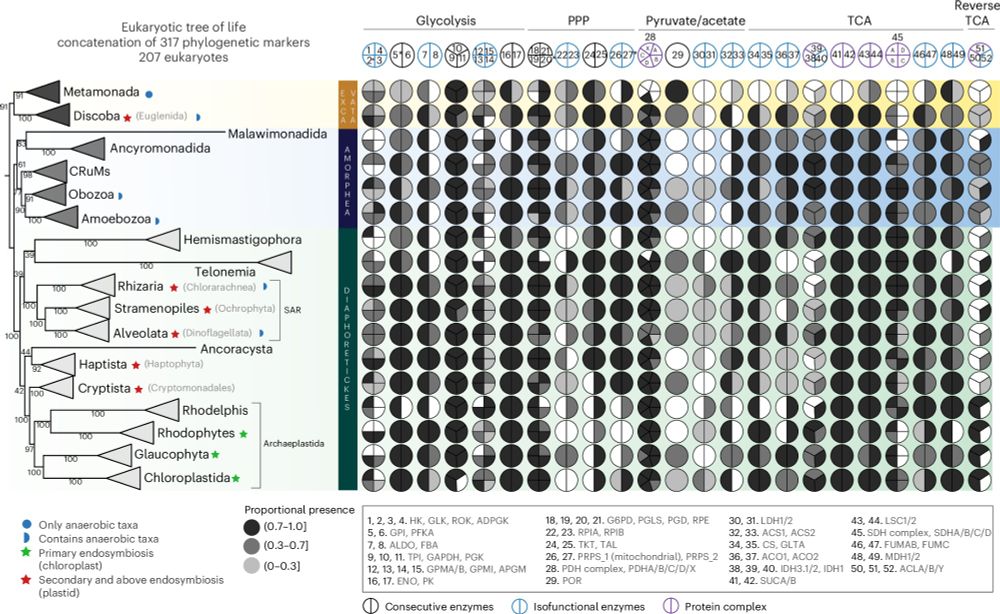
A robustly rooted tree of eukaryotes reveals their excavate ancestry - Nature
The root of the eukaryote Tree of Life is estimated from a new, larger dataset of mitochondrial proteins including all known eukaryotic supergroups, showing it lies between two multi-supergroup assemb...
www.nature.com
Tom Williams
@tweethinking.bsky.social
· Feb 18
Tom Williams
@tweethinking.bsky.social
· Feb 17
Tom Williams
@tweethinking.bsky.social
· Feb 17
Tom Williams
@tweethinking.bsky.social
· Feb 17

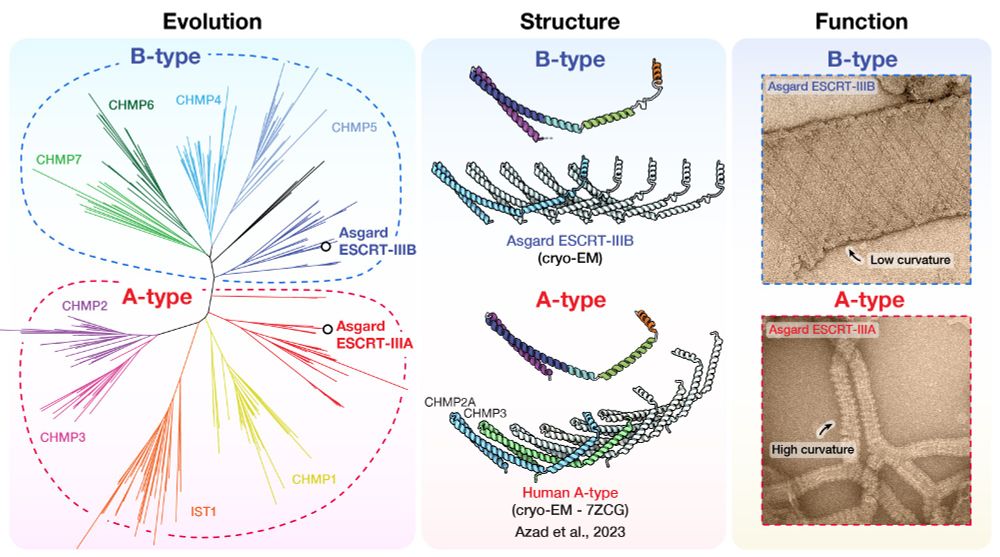
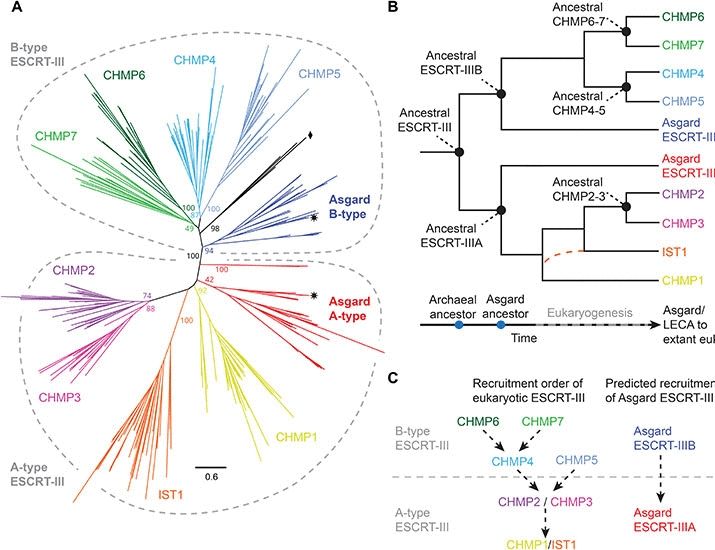
Phylogenomic analyses reveal that Panguiarchaeum is a clade of genome-reduced Asgard archaea
The Asgard archaea are a diverse archaeal phylum that includes the host lineage from which eukaryotes evolved. Due to the importance of the Asgard archaea for our understanding of cellular evolution a...
www.biorxiv.org
Reposted by Tom Williams
Reposted by Tom Williams