Fyodor Urnov
@urnov.bsky.social
2.2K followers
600 following
370 posts
Clinic-minded genome + epigenome editor. Professor of Molecular Therapeutics, UC Berkeley. Director for Technology and Translation, Innovative Genomics Institute, ibid.
Beatles fan.
Art work credit: Fyodor Vasiliev "A Meadow in the Rain" (1872).
Posts
Media
Videos
Starter Packs
Fyodor Urnov
@urnov.bsky.social
· Sep 7

Potent and uniform fetal hemoglobin induction via base editing - Nature Genetics
A comparison of fetal hemoglobin gene editing strategies using human sickle cell disease donor cells and in vivo transplantation finds that adenine base editing of the –175A>G site in the γ-globin ...
www.nature.com
Fyodor Urnov
@urnov.bsky.social
· Sep 7
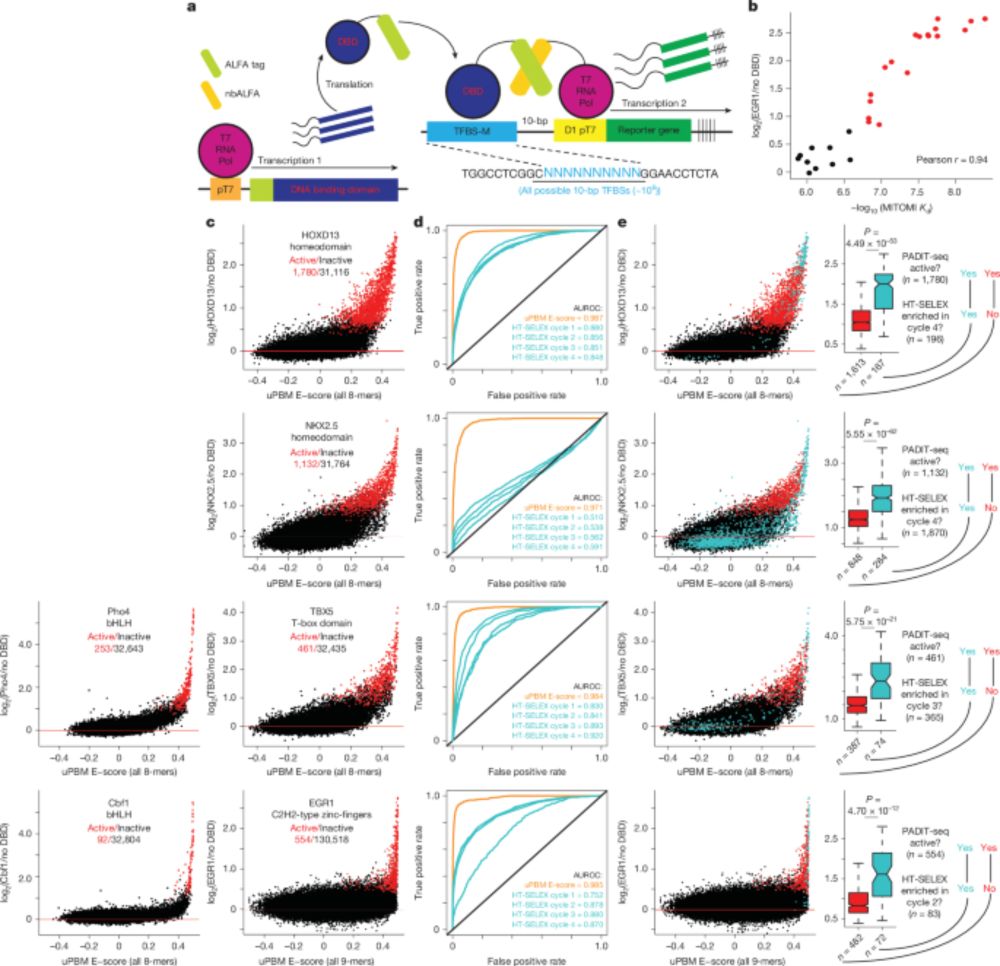
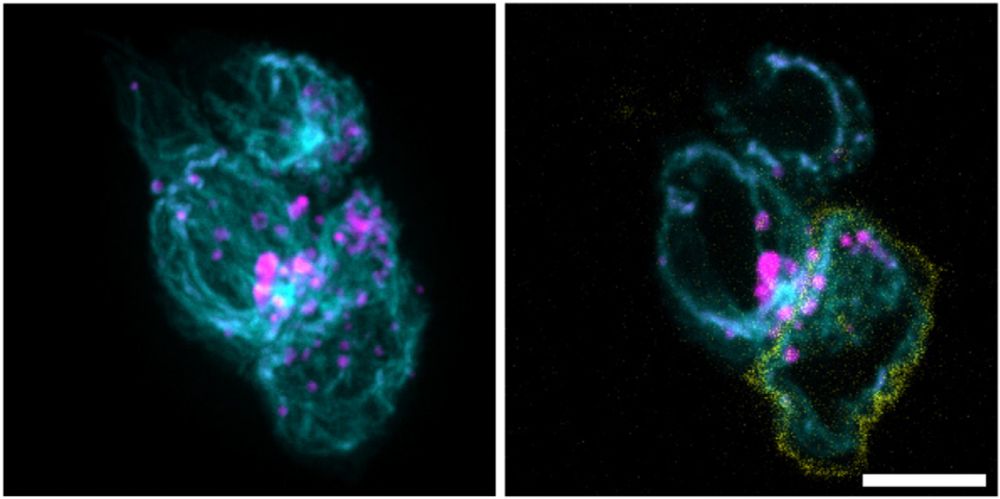
Multiple overlapping binding sites determine transcription factor occupancy - Nature
A new method enables comprehensive screening and identification of low-affinity DNA binding sites for transcription factors, and reveals that nucleotides flanking high-affinity binding sites create ov...
www.nature.com
Fyodor Urnov
@urnov.bsky.social
· Sep 1
DNA Methylation Ageing Atlas Across 17 Human Tissues
Aging involves widespread epigenetic remodeling across tissues, yet the nature and consistency of these changes remain unclear. We conducted a meta-analysis of more than 15,000 human methylomes spanni...
www.researchsquare.com
Fyodor Urnov
@urnov.bsky.social
· Aug 13

A dogged reporter takes on a mysterious cabal in 'The Diary of Lies'
Philip Miller's sinister thriller is set in a Great Britain that's lost its bearings. But even when she's terrified, fictional journalist Shona Sandison will always risk everything to get the story.
www.npr.org
Fyodor Urnov
@urnov.bsky.social
· Aug 6
Fyodor Urnov
@urnov.bsky.social
· Aug 5
Reposted by Fyodor Urnov
Ryan Cross
@scienceboss.bsky.social
· Jul 9

'CRISPR Cures' center launches with $20M to make customized gene editing therapies
Scientists seeking to make personalized gene editing therapies a reality for more people have launched a new initiative to develop cures for the rarest of diseases left behind by drug companies.
endpoints.news
Reposted by Fyodor Urnov
Fyodor Urnov
@urnov.bsky.social
· Jun 10
Jackson Hoffman
@jxhoffman.bsky.social
· Jun 10

Pioneering new enhancers by GATA3: role of facilitating transcription factors and chromatin remodeling
Abstract. Pioneer transcription factors (PTFs) bind to inaccessible chromatin and recruit collaborating transcription factors to promote chromatin accessib
doi.org
Fyodor Urnov
@urnov.bsky.social
· Jun 10
Fyodor Urnov
@urnov.bsky.social
· Jun 9
Fyodor Urnov
@urnov.bsky.social
· Jun 9
Fyodor Urnov
@urnov.bsky.social
· Jun 9
Fyodor Urnov
@urnov.bsky.social
· Jun 9
Fyodor Urnov
@urnov.bsky.social
· Jun 9
Fyodor Urnov
@urnov.bsky.social
· Jun 8
Fyodor Urnov
@urnov.bsky.social
· Jun 8
Fyodor Urnov
@urnov.bsky.social
· Jun 7