Veesler lab
@veeslerlab.bsky.social
990 followers
360 following
11 posts
Pathogen entry into cells, host immune responses & vaccine design: structural biology, protein design, virology & immunology @hhmi.bsky.social
@uofwa.bsky.social
Posts
Media
Videos
Starter Packs
Reposted by Veesler lab
Reposted by Veesler lab
Reposted by Veesler lab
Reposted by Veesler lab
Reposted by Veesler lab
Reposted by Veesler lab
cryoEM papers
@cryoempapers.bsky.social
· Jul 11
Reposted by Veesler lab
Veesler lab
@veeslerlab.bsky.social
· May 18

Potent neutralization of Marburg virus by a vaccine-elicited monoclonal antibody
Marburg virus (MARV) is a filovirus that causes a severe and often lethal hemorrhagic fever. Despite the increasing frequency of MARV outbreaks, no vaccines or therapeutics are licensed for use in hum...
www.biorxiv.org
Veesler lab
@veeslerlab.bsky.social
· May 14

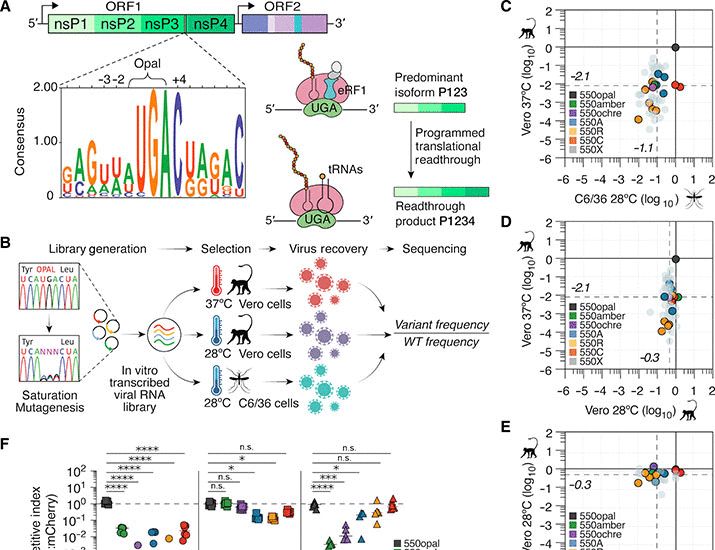
SARS-CoV-2 nsp1 mediates broad inhibition of translation in mammals
Gen et al. demonstrate that SARS-CoV-2 nsp1 potently blocks translation in Rhinolophus
lepidus bat cells and several other mammalian cell lines, concurring with the broad
species tropism of SARS-CoV-2...
www.cell.com
Veesler lab
@veeslerlab.bsky.social
· May 14
Veesler lab
@veeslerlab.bsky.social
· May 14

Phylogeny-driven design of broadly protective sarbecovirus receptor-binding domain nanoparticle vaccines
Vaccines against emerging SARS-CoV-2 variants and sarbecoviruses with pandemic potential must elicit a robust humoral immune response in a population imprinted with the SARS-CoV-2 spike (S) protein. H...
www.biorxiv.org
Reposted by Veesler lab
Reposted by Veesler lab
Reposted by Veesler lab
Reposted by Veesler lab
Reposted by Veesler lab
Reichow Lab
@reichowlab.bsky.social
· Feb 21
Veesler lab
@veeslerlab.bsky.social
· Feb 21

ACE2 utilization of HKU25 clade MERS-related coronaviruses with broad geographic distribution
Dipeptidyl peptidase-4 (DPP4) is a well-established receptor for several MERS-related coronaviruses (MERSr-CoVs) isolated from humans, camels, pangolins, and bats. However, the receptor usage of many ...
www.biorxiv.org
Reposted by Veesler lab
Veesler lab
@veeslerlab.bsky.social
· Feb 7
Veesler lab
@veeslerlab.bsky.social
· Feb 7

Multiple independent acquisitions of ACE2 usage in MERS-related coronaviruses
Two coronaviruses related to MERS-CoV, which infect European Pipistrellus bats, have
been found to use ACE2 as their receptor. These viruses target an ACE2 surface region
distant from those recognized...
www.cell.com
Veesler lab
@veeslerlab.bsky.social
· Feb 7

Molecular basis of convergent evolution of ACE2 receptor utilization among HKU5 coronaviruses
The family of coronaviruses known as merbecoviruses has generally been thought to
use the host cell protein DPP4 for entry, but this study shows that the merbecovirus
HKU5 can enter host cells of the ...
www.cell.com

![A lot of what NIH funds is basic science, which focuses on discoveries that might or might not result in practical applications. How can you describe the value of basic science to the public? To most people, it seems nebulous. It might take 30 years of research to pay off in a practical way.
Yes, it’s hard for people to understand the importance of basic science. I think the best way to deal with that is to talk about examples.
I’ll reflect on my experience with cystic fibrosis research. The effort to try to do something for that almost-uniformly fatal disease started as a basic science enterprise to try to discover the nature of the gene that is misspelled. That was a hard slog back in the 1980s. Nobody would have supported [research into it] except NIH.
That work by my University of Michigan lab, working with Lap-Chee Tsui at the Hospital for Sick Kids in Toronto, led to the discovery of the gene [and the mutation that causes the disease] in 1989. Then, a partnership of NIH-funded research, philanthropy from the Cystic Fibrosis Foundation, and investments by biotech and pharma led to a therapeutic breakthrough.
Now, 30 years after the gene discovery, young people with cystic fibrosis are planning for retirement instead of an early funeral.](https://cdn.bsky.app/img/feed_thumbnail/plain/did:plc:7e6gdj33irtt6i7vc6hozfah/bafkreic6ydv2aqftxi3caocigytazotzicudx4uww37l2acn3misi6dkka@jpeg)