William Garland
@willgarland.bsky.social
390 followers
400 following
29 posts
RNA decay, RNA sorting, transcriptional regulation
Aarhus University, Denmark 🇩🇰
Posts
Media
Videos
Starter Packs
William Garland
@willgarland.bsky.social
· Jul 22
Torben Heick Jensen
@heick.bsky.social
· Jun 27

A postdoc position in the Torben Heick Jensen lab, Aarhus University, Denmark: Mammalian Nuclear RNA Production and Turnover Systems - Vacancy at Aarhus University
Vacancy at Department of Molecular Biology and Genetics - RNA Biology and Innovation, Aarhus University
mbg.au.dk
Reposted by William Garland
Torben Heick Jensen
@heick.bsky.social
· Jun 27

A postdoc position in the Torben Heick Jensen lab, Aarhus University, Denmark: Mammalian Nuclear RNA Production and Turnover Systems - Vacancy at Aarhus University
Vacancy at Department of Molecular Biology and Genetics - RNA Biology and Innovation, Aarhus University
mbg.au.dk
William Garland
@willgarland.bsky.social
· Jun 17

International recognition for Aarhus researchers: RNA science takes off
RNA research in Aarhus is taking a significant leap forward. Two researchers from the same department and research environment have both just been awarded one of Europe’s most prestigious research gra...
mbg.au.dk
William Garland
@willgarland.bsky.social
· Apr 28
Reposted by William Garland
Reposted by William Garland
Torben Heick Jensen
@heick.bsky.social
· Feb 20
Reposted by William Garland
Reposted by William Garland
Leopold Parts
@leopoldparts.bsky.social
· Jan 31
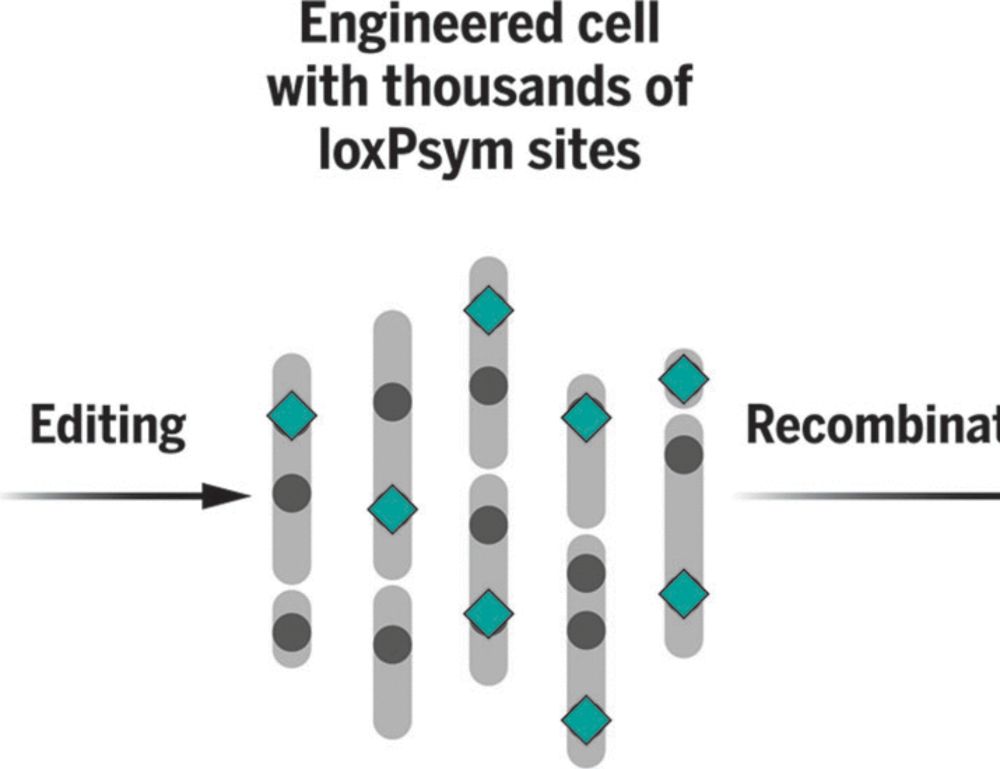
Randomizing the human genome by engineering recombination between repeat elements
We lack tools to edit DNA sequences at scales necessary to study 99% of the human genome that is noncoding. To address this gap, we applied CRISPR prime editing to insert recombination handles into re...
www.science.org
Reposted by William Garland
Reposted by William Garland
Cell Press
@cellpress.bsky.social
· Jan 31
Reposted by William Garland
Reposted by William Garland
William Garland
@willgarland.bsky.social
· Dec 21
William Garland
@willgarland.bsky.social
· Dec 13

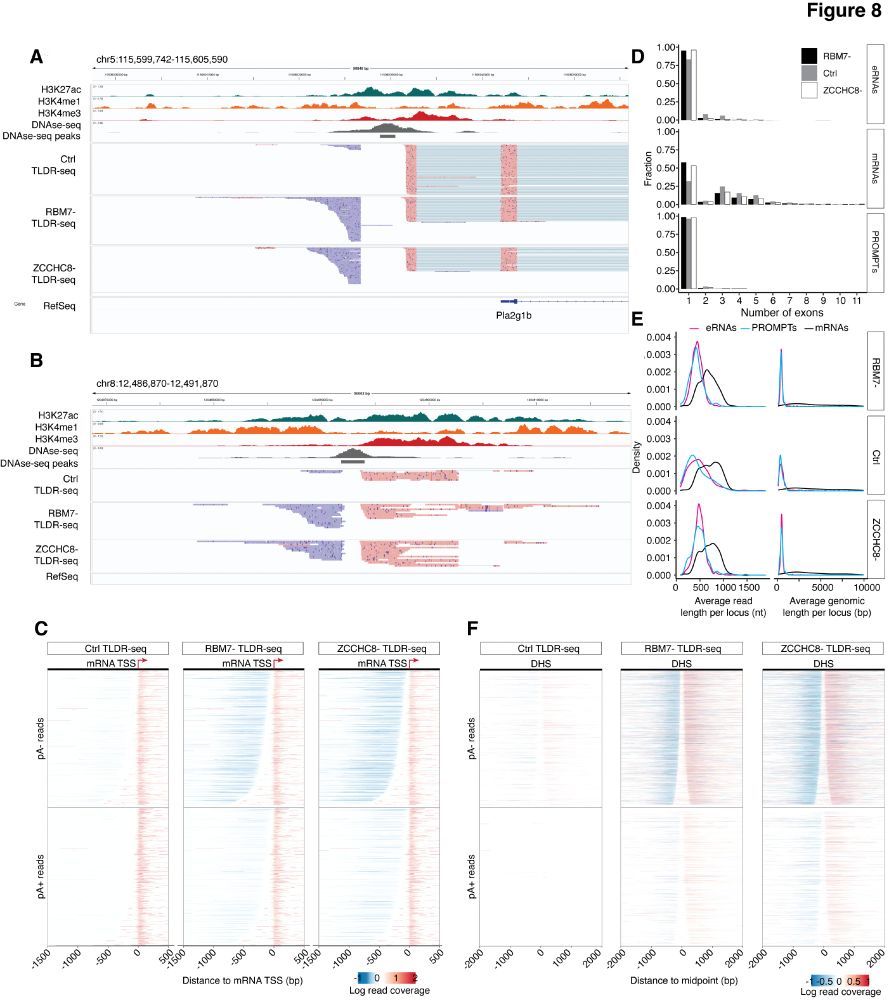
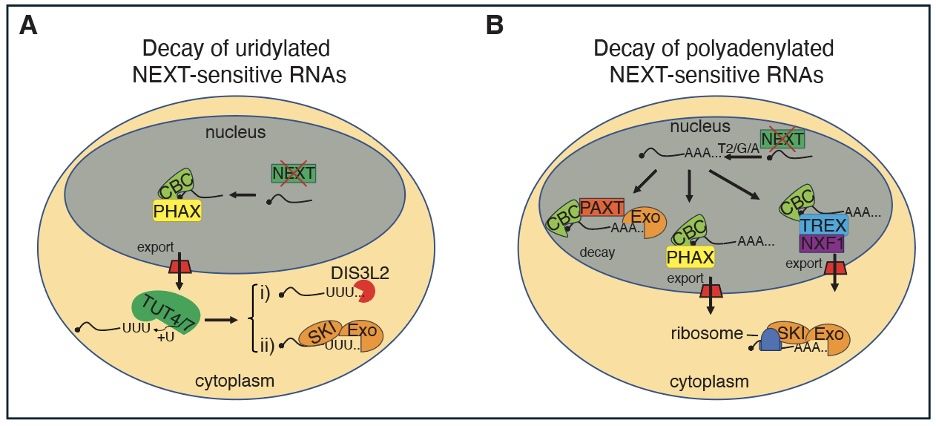
RNA 3′end tailing safeguards cells against products of pervasive transcription termination - Nature Communications
Pervasive transcription generates numerous unadenylated RNAs, usually degraded by the NEXT/nuclear exosome pathway. Here, the authors show that, upon NEXT inactivation, these RNAs are removed by compe...
doi.org