Yizhou Hu(Eugene)
@yizhoueugene.bsky.social
43 followers
110 following
14 posts
Researcher;System Medicine,developmental plasticity in nervous system cancers, psoriatic skin ;@Karolinska Inst. , @Wihuri Inst. & Univ. of Helsinki
Posts
Media
Videos
Starter Packs
Pinned
Reposted by Yizhou Hu(Eugene)
Reposted by Yizhou Hu(Eugene)
Reposted by Yizhou Hu(Eugene)
Igor Adameyko
@adameykolab.bsky.social
· May 20
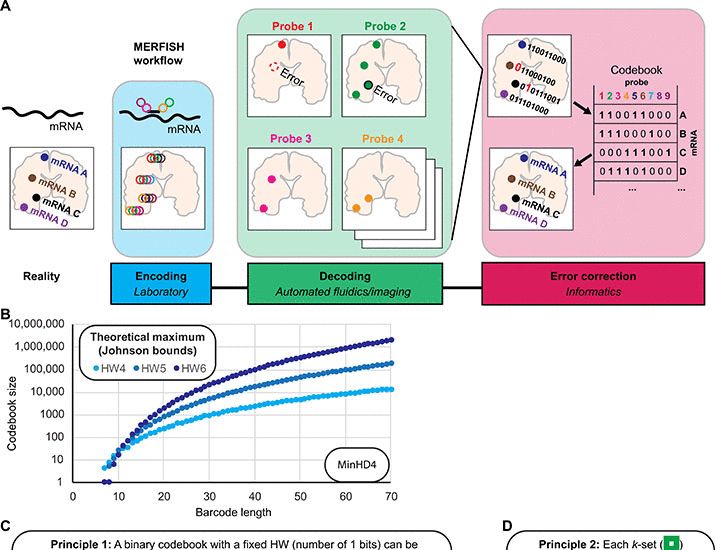
Boosting multiplexing capabilities for error-robust spatial transcriptomic methods using a set exchange approach
Using an algorithm, highly optimized error-robust codebooks for spatial transcriptomic methods can systematically be produced.
www.science.org
Reposted by Yizhou Hu(Eugene)
Reposted by Yizhou Hu(Eugene)
Reposted by Yizhou Hu(Eugene)
Patrik Ernfors
@patrikernfors.bsky.social
· Mar 24
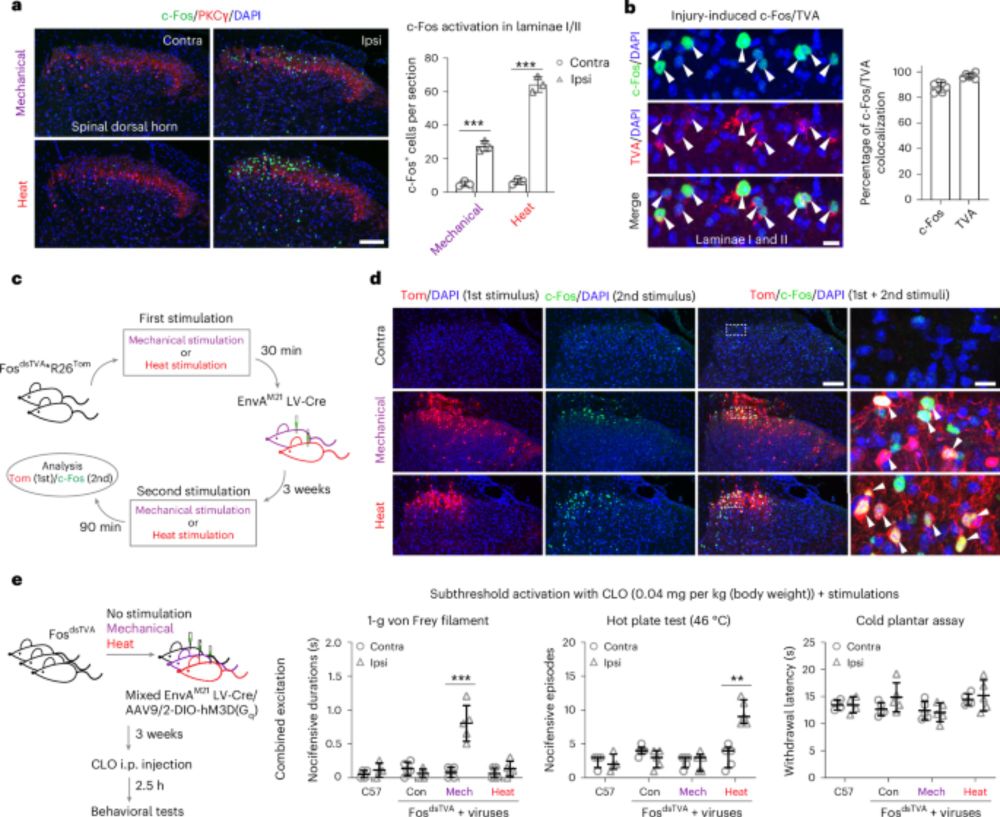
Neural ensembles that encode nocifensive mechanical and heat pain in mouse spinal cord - Nature Neuroscience
Zhang et al. identify unimodal neural representations in the spinal cord of cutaneous mechanical and heat stimuli gated by a shared feed-forward local inhibitory neuron type and a neural transition du...
www.nature.com