Congrats Hannah Ochner and authors on this important paper! Strong collaboration with @kiranrpatil.bsky.social
www.biorxiv.org/cgi/content/...
@mrclmb.bsky.social @wellcometrust.bsky.social

Reposted by: Kiran Raosaheb Patil
#science #microbiome #health
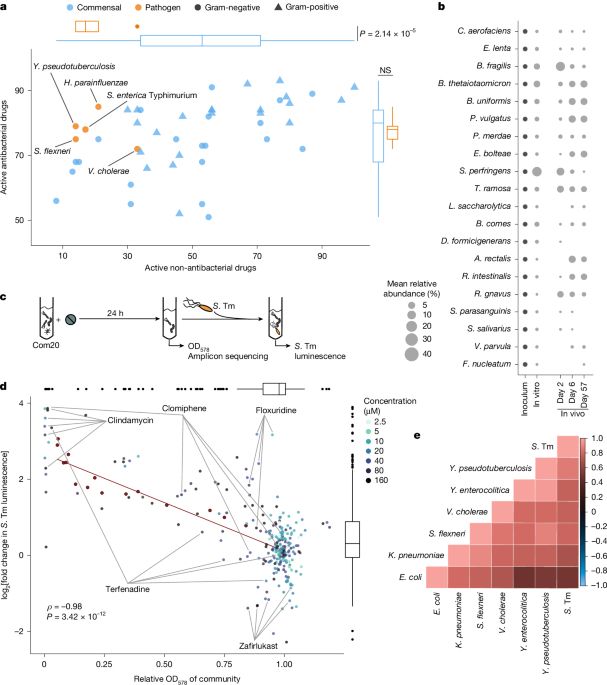
www.nature.com/articles/s41...

Reposted by: Kiran Raosaheb Patil
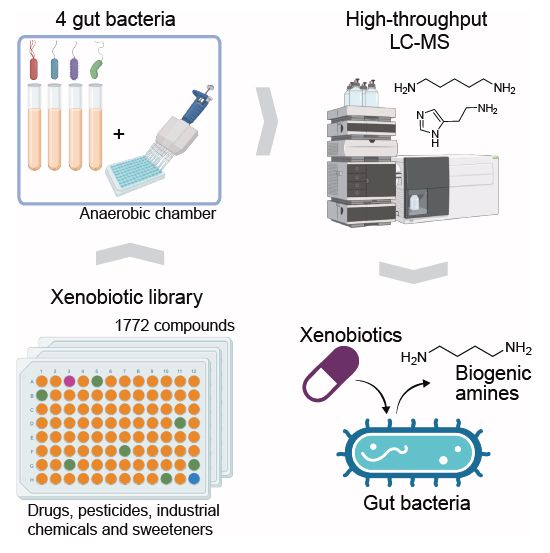





www.nature.com/articles/s41...
Reposted by: Kiran Raosaheb Patil
🚨Out now!
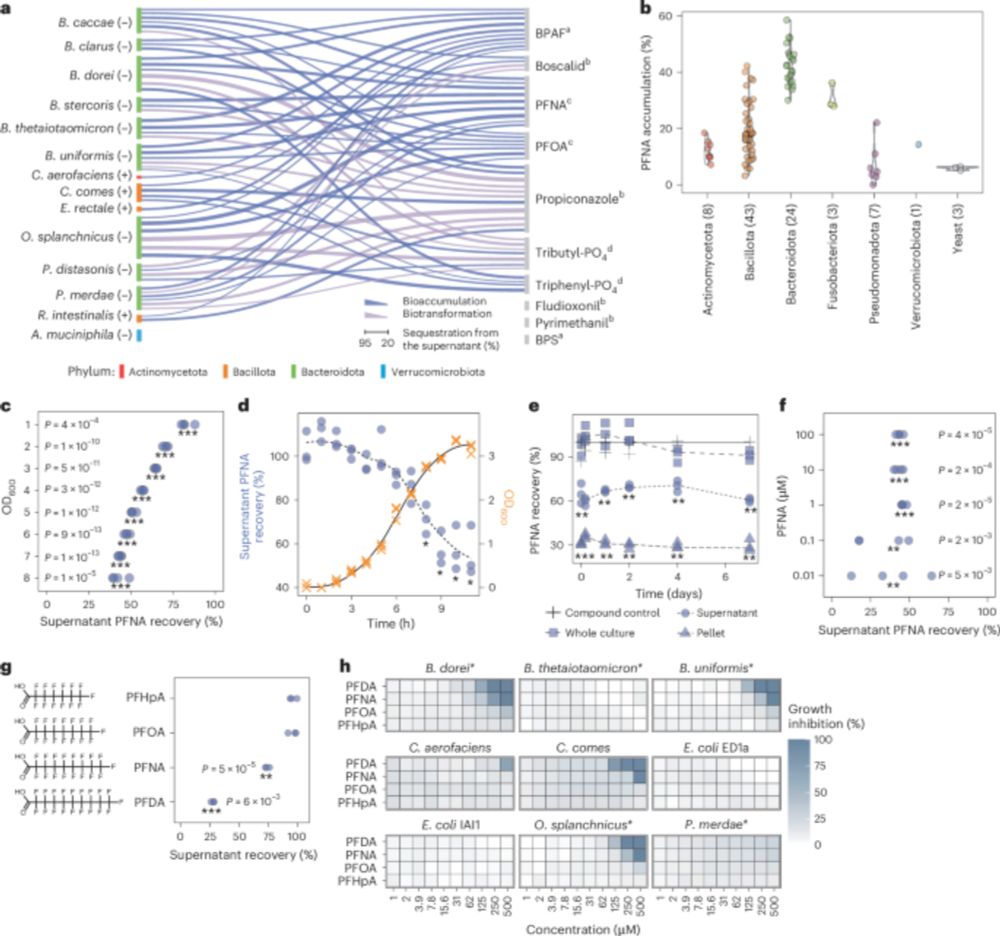
Human gut bacteria bioaccumulate forever chemicals, in intracellular aggregates and colonization of gnotobiotic mice with these bioaccumulating bacteria increases faecal PFAS excretion. @kiranrpatil.bsky.social
#MicroSky #MicrobiomeSky 🦠
www.nature.com/articles/s41...
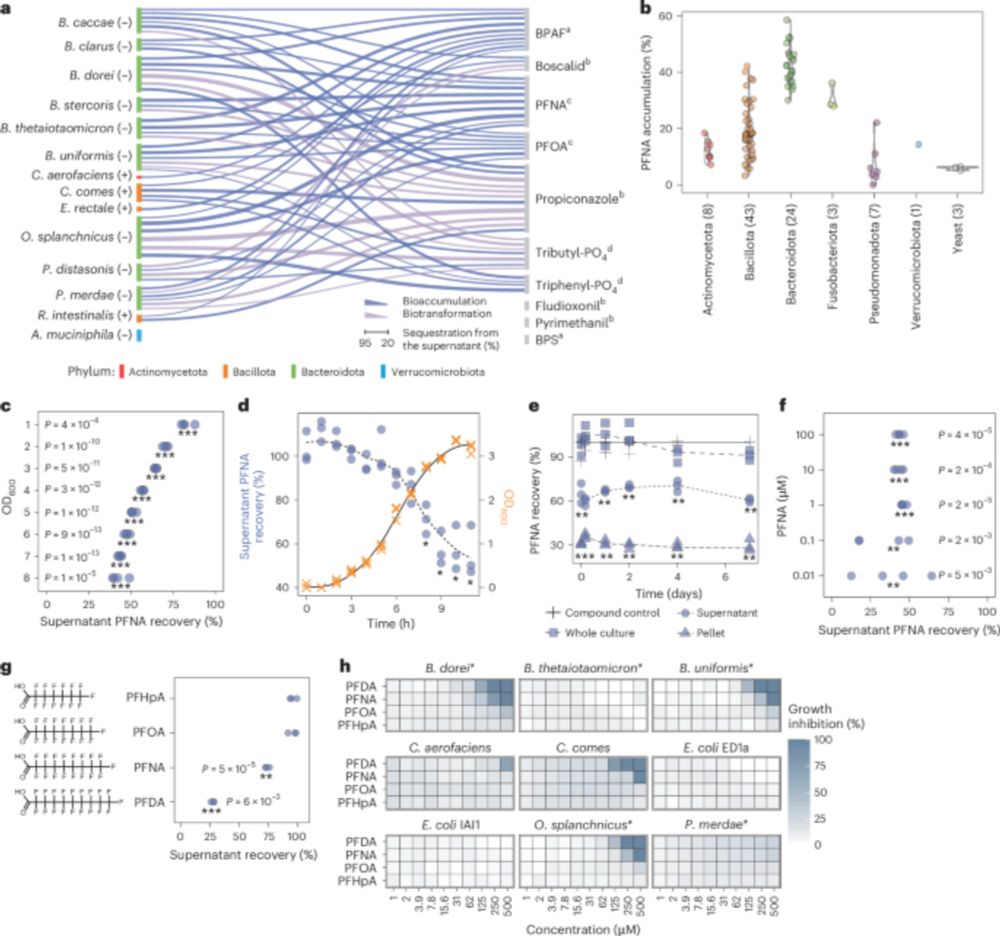

Reposted by: Kiran Raosaheb Patil
Reposted by: Kiran Raosaheb Patil
#Cambridgeshire #BBCNews
🔗: https://www.bbc.com/news/arti...

Reposted by: Kiran Raosaheb Patil, Michael Wagner
They can shield us from harmful chemicals—and the latest study from Kiran Patil's lab (@kiranrpatil.bsky.social)
uncovers how. We’re proud to have contributed to this exciting work!
#Microbiome #Detox
www.nature.com/articles/s41...
Reposted by: Kiran Raosaheb Patil
Read more here: buff.ly/4FsVFsS
@kiranrpatil.bsky.social @indraroux.bsky.social
Reposted by: Kiran Raosaheb Patil
Find out more here: buff.ly/5akzsgs
@kiranrpatil.bsky.social @skamrad.bsky.social
Reposted by: Kiran Raosaheb Patil
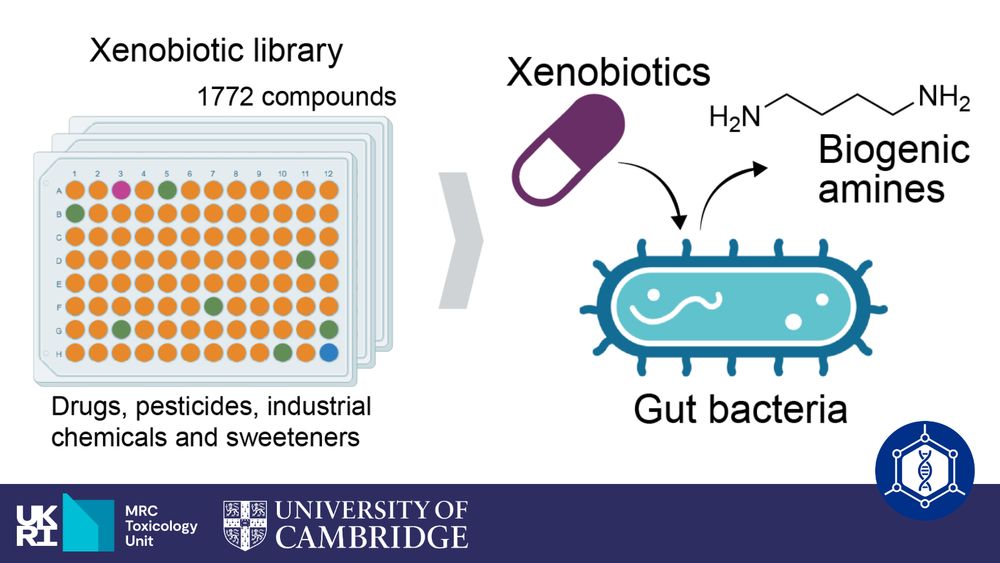
We systematically map proteomic and metabolomic interactions between >100 pairs of gut bacteria, detecting metabolic interactions and functionally-related clusters of proteins.
www.biorxiv.org/content/10.1...
@kiranrpatil.bsky.social

Reposted by: Kiran Raosaheb Patil
We show how glycolytic activity instructs germ layer proportions through regulation of Nodal and Wnt signaling - happy to finally share this 😊
doi.org/10.1016/j.st...
B2B with @jesseveenvliet.bsky.social lab: doi.org/10.1016/j.st... 🤩


Reposted by: Kiran Raosaheb Patil
@kiranrpatil.bsky.social:
Obligate cross-feeding of metabolites is common in soil microbial communities
By Ghada Yousif @metagenomez.bsky.social with @swagatika.bsky.social @isamirgiri.bsky.social Sharvari Harshe et al.
www.biorxiv.org/content/10.1...
🧵👇

