@ecol-evol.bsky.social was featured in @science.org #ScienceShots written by
@sahasmehra.bsky.social
#Cyclosa #decoy #spiders #evolution #Peru #Philippines

@ecol-evol.bsky.social was featured in @science.org #ScienceShots written by
@sahasmehra.bsky.social
#Cyclosa #decoy #spiders #evolution #Peru #Philippines
Cuyabeno Reserve, Ecuador 2022

Cuyabeno Reserve, Ecuador 2022
www.nature.com/articles/d41...

www.nature.com/articles/d41...

Meet CLAE: a high-fidelity Nanopore sequencing strategy that pushes accuracy to Q30, boosts throughput >800 Mb/100 pores, and recovers full-length viral genomes from complex samples 🌊🦠
👉 [https://doi.org/10.1002/advs.202505978]
#viromics #nanopore #genomics
www.medrxiv.org/content/10.1...
www.medrxiv.org/content/10.1...
Please RePost 🙏
We just released reference genomes for another 250 strains to the ATCC Genome Portal (genomes.ATCC.org)! We now have assemblies for
_5,750 microbial genomes_
AND
all assemblies w/ @nanoporetech.com data will include DNA methylation profiles too (details👇)
#genomics 1/
Please RePost 🙏
We just released reference genomes for another 250 strains to the ATCC Genome Portal (genomes.ATCC.org)! We now have assemblies for
_5,750 microbial genomes_
AND
all assemblies w/ @nanoporetech.com data will include DNA methylation profiles too (details👇)
#genomics 1/
Join Robert James, Angela Hickey, and Jose Alexander as they share how real-time, Oxford Nanopore sequencing is transforming microbiology—from single bacterial isolates to the complexities of the microbiome.
nanoporetech.com/about/events...

Join Robert James, Angela Hickey, and Jose Alexander as they share how real-time, Oxford Nanopore sequencing is transforming microbiology—from single bacterial isolates to the complexities of the microbiome.
nanoporetech.com/about/events...
wwwnc.cdc.gov/eid/article/...







myloasm-docs.github.io


First, we saw how the technology continued to advance research in 2024 with ~2/3 of ONT publications in micro/infectious #nanoporeconf
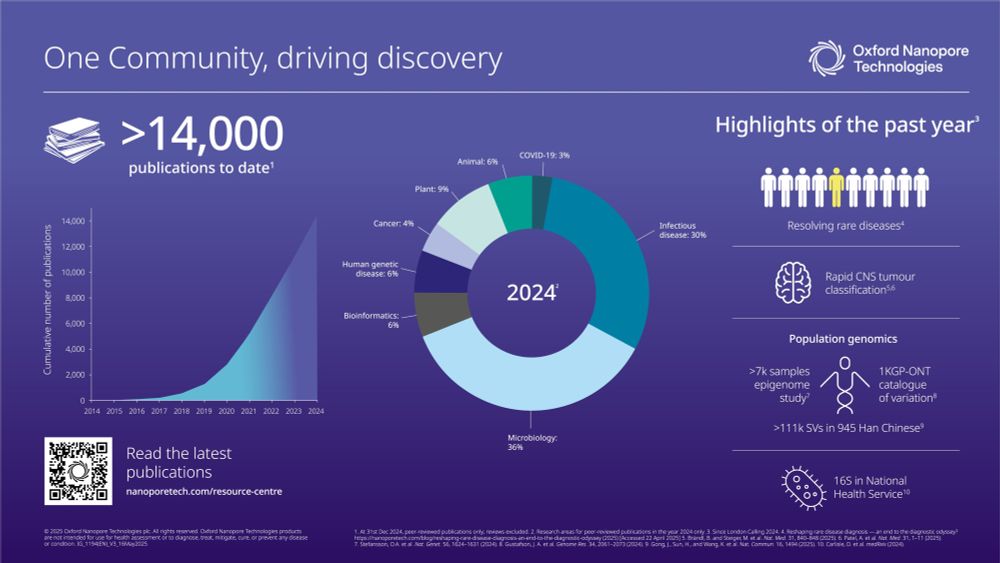
First, we saw how the technology continued to advance research in 2024 with ~2/3 of ONT publications in micro/infectious #nanoporeconf
Who’s going to be working their tails off trying to get this data out before ASM Microbe? We are. 🙃
Stay tuned! Major updates coming to the ATCC Genome Portal starting w/methylation data on 1000’s of microbes
Updating the ATCC Genome Portal to be able to deliver the DNA methylation data we have on over 5,000 bacteria and fungi…
We just finished rebasecalling all of the raw @nanoporetech.com data we’ve produced since 2019 with the latest version of Dorado.🙃
Updating the ATCC Genome Portal to be able to deliver the DNA methylation data we have on over 5,000 bacteria and fungi…
We just finished rebasecalling all of the raw @nanoporetech.com data we’ve produced since 2019 with the latest version of Dorado.🙃
www.biorxiv.org/content/10.1...
www.biorxiv.org/content/10.1...



