Interested in the functional annotation of genomic and metagenomic data, carbohydrate metabolism, #bifidobacteria
www.biorxiv.org/content/10.1...

www.biorxiv.org/content/10.1...
www.nature.com/articles/s41...

www.nature.com/articles/s41...










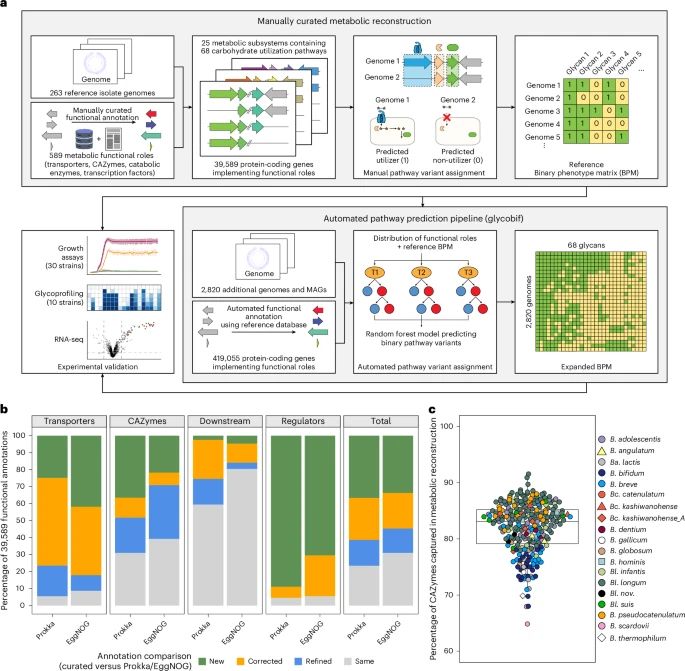
Key takeaways:
🔹 Host ancestry + diet shape Bif evolution
🔹 Mammals enriched for carb-busting enzymes
🔹 Untapped diversity in non-human hosts = new probiotic potential
@halllab.bsky.social survey insects, reptiles, birds & mammals to uncover how Bifidobacterium adapts to hosts. Bifidobacterium & host exhibit strong co-phylogenetic associations, driven by vertical transmission & dietary selection
www.cell.com/cell-host-mi...

Key takeaways:
🔹 Host ancestry + diet shape Bif evolution
🔹 Mammals enriched for carb-busting enzymes
🔹 Untapped diversity in non-human hosts = new probiotic potential
We finally have the ability to study microbial activity on skin and identify key functional genes playing a role in diseases.
Amazing team of Chia Minghao and Amanda Ng 👏
nature.com/articles/s41...
We finally have the ability to study microbial activity on skin and identify key functional genes playing a role in diseases.
Amazing team of Chia Minghao and Amanda Ng 👏
nature.com/articles/s41...
journals.asm.org/doi/full/10....

journals.asm.org/doi/full/10....
academic.oup.com/bioinformati...

academic.oup.com/bioinformati...
www.jbc.org/article/S002...

www.jbc.org/article/S002...
www.jbc.org/article/S002...

www.jbc.org/article/S002...
link.springer.com/article/10.1...

link.springer.com/article/10.1...

