IMO, ICPC, Xoogler, Rust, road-cycling, hiking, wild camping, photography

Currently we have .gitlab, .config, .cargo, ..., and it creates clutter.

I’d immediately get rid of the “documents” thing and anchors and many of the strings.

Then a year at 100m off the Thames, continued by 8 years on a tributary of the Rhine.
Now moving to mid-Rhine, but sadly 5km away from it.
So yeah, I'm a Rhine boy :)
Mine is the Patuxent.
Then a year at 100m off the Thames, continued by 8 years on a tributary of the Rhine.
Now moving to mid-Rhine, but sadly 5km away from it.
So yeah, I'm a Rhine boy :)
Alignoth generates self-contained interactive HTML read alignment plots from BAM files – Rust-based, portable, and ideal for headless workflows.
📄 doi.org/10.1093/bioi...
#bioinformatics #genomics #rust @johanneskoester.bsky.social

Alignoth generates self-contained interactive HTML read alignment plots from BAM files – Rust-based, portable, and ideal for headless workflows.
📄 doi.org/10.1093/bioi...
#bioinformatics #genomics #rust @johanneskoester.bsky.social
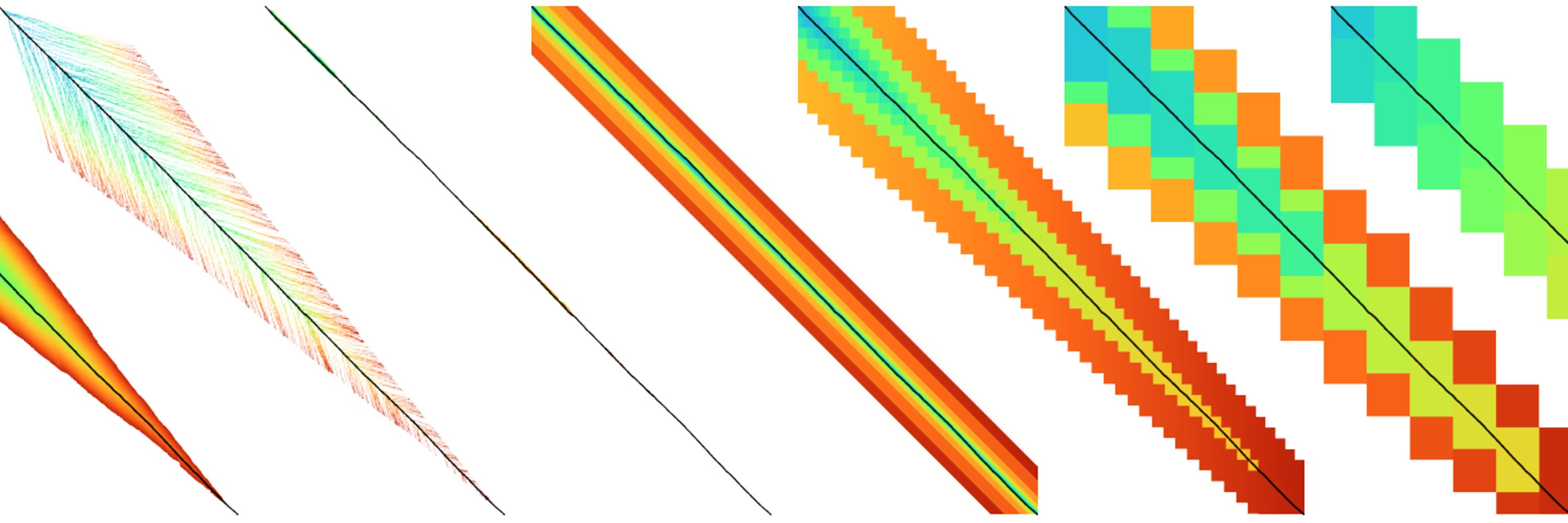
60% + 0.18% = 66.85% of n, which is not so negligible.
- Gonzales, Grabowski, Makinen, Navarro
60% + 0.18% = 66.85% of n, which is not so negligible.
- Gonzales, Grabowski, Makinen, Navarro
@recombseq.bsky.social @pashadag.bsky.social @ksahlin.bsky.social
@recombseq.bsky.social @pashadag.bsky.social @ksahlin.bsky.social
(The footnote goes to Rank9 and Dijkstra)

(The footnote goes to Rank9 and Dijkstra)






