🔗 doi.org/10.1101/2025...
💻 github.com/Bitbol-Lab/r...
1/7

We introduce RAG-ESM, a retrieval-augmented framework that makes pretrained protein language models (like ESM2) homology-aware with minimal training cost.
📄 Paper: journals.aps.org/prxlife/abst...
We developed salad (sparse all-atom denoising), a family of blazing fast protein structure diffusion models.
Paper: nature.com/articles/s42256-…
Code: github.com/mjendrusch/salad
Data: zenodo.org/records/14711580
1/🧵
Salad is significantly faster than comparable methods, and designing proteins that don't exist in nature can have applications in many scientific fields.
www.nature.com/articles/s42...

We developed salad (sparse all-atom denoising), a family of blazing fast protein structure diffusion models.
Paper: nature.com/articles/s42256-…
Code: github.com/mjendrusch/salad
Data: zenodo.org/records/14711580
1/🧵
- AI Scientist 👉 lnkd.in/eDXHH4E8
- AI Scientist, Drug Creation 👉 lnkd.in/eEvGyaTR
You'll work on antibody sequence/structure design, antibody-antigen co-folding, antibody-antigen binding prediction, physics-based methodologies, and more!
DMs welcome!
- AI Scientist 👉 lnkd.in/eDXHH4E8
- AI Scientist, Drug Creation 👉 lnkd.in/eEvGyaTR
You'll work on antibody sequence/structure design, antibody-antigen co-folding, antibody-antigen binding prediction, physics-based methodologies, and more!
DMs welcome!
We introduce RAG-ESM, a retrieval-augmented framework that makes pretrained protein language models (like ESM2) homology-aware with minimal training cost.
📄 Paper: journals.aps.org/prxlife/abst...
🔗 doi.org/10.1101/2025...
💻 github.com/Bitbol-Lab/r...
1/7

We introduce RAG-ESM, a retrieval-augmented framework that makes pretrained protein language models (like ESM2) homology-aware with minimal training cost.
📄 Paper: journals.aps.org/prxlife/abst...
🔗 www.biorxiv.org/content/10.1...

🔗 www.biorxiv.org/content/10.1...
doi.org/10.1093/bioi...

doi.org/10.1093/bioi...
Last Friday I defended my thesis titled: "Revealing and Exploiting Coevolution through Protein Language Models".
It was an amazing journey where I met some incredible people. Thank you all ❤️

Last Friday I defended my thesis titled: "Revealing and Exploiting Coevolution through Protein Language Models".
It was an amazing journey where I met some incredible people. Thank you all ❤️
By modulating SWI/SNF remodeling at ancient transposable elements - LINE/L2s and SINE/MIRs, a "noncanonical" KZFP called ZNF436 protects cardiomyocytes from losing their identity.
🫀heartbeat on 🔁 repeat
www.biorxiv.org/content/10.1... #TEsky
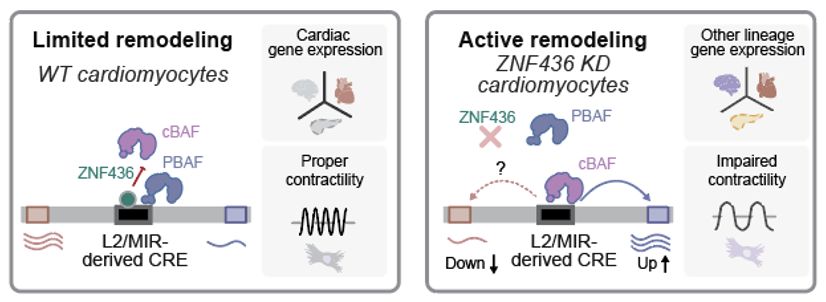
By modulating SWI/SNF remodeling at ancient transposable elements - LINE/L2s and SINE/MIRs, a "noncanonical" KZFP called ZNF436 protects cardiomyocytes from losing their identity.
🫀heartbeat on 🔁 repeat
www.biorxiv.org/content/10.1... #TEsky
🔗 doi.org/10.1101/2025...
💻 github.com/Bitbol-Lab/r...
1/7

🔗 doi.org/10.1101/2025...
💻 github.com/Bitbol-Lab/r...
1/7
#ICLR2025 (Oral Presentation)
🔥 Project page: research.nvidia.com/labs/genair/...
📜 Paper: arxiv.org/abs/2503.00710
🛠️ Code and weights: github.com/NVIDIA-Digit...
🧵Details in thread...
(1/n)
#ICLR2025 (Oral Presentation)
🔥 Project page: research.nvidia.com/labs/genair/...
📜 Paper: arxiv.org/abs/2503.00710
🛠️ Code and weights: github.com/NVIDIA-Digit...
🧵Details in thread...
(1/n)
We trained 2 new models. Like BERT, but modern. ModernBERT.
Not some hypey GenAI thing, but a proper workhorse model, for retrieval, classification, etc. Real practical stuff.
It's much faster, more accurate, longer context, and more useful. 🧵
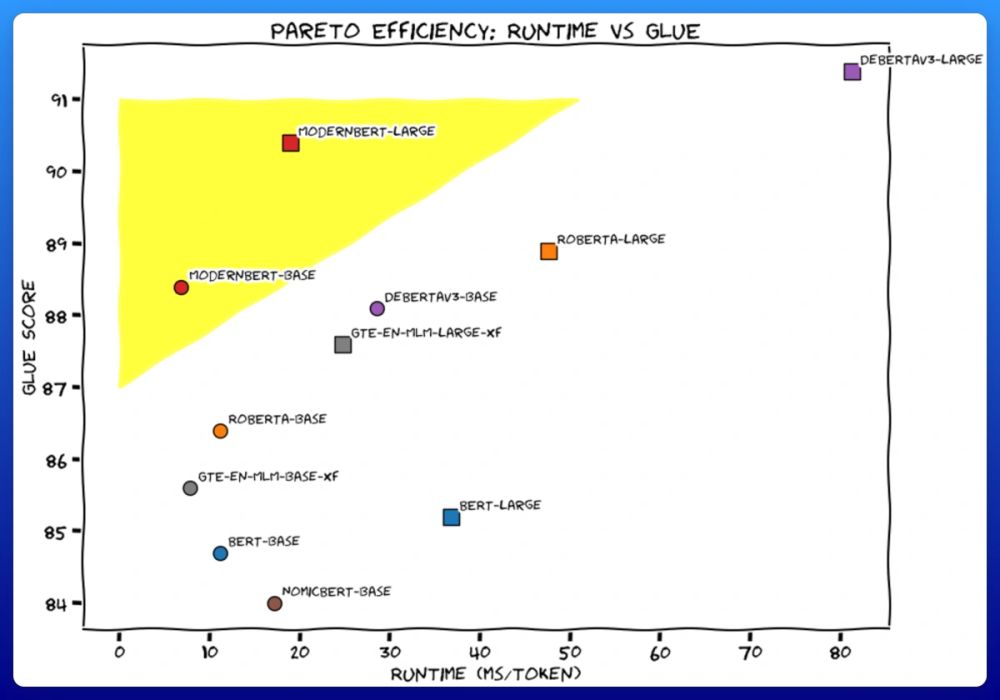
We trained 2 new models. Like BERT, but modern. ModernBERT.
Not some hypey GenAI thing, but a proper workhorse model, for retrieval, classification, etc. Real practical stuff.
It's much faster, more accurate, longer context, and more useful. 🧵
Thanks a lot to the great co-authors! Esp
@rne.bsky.social & Yuanqi Du.

Thanks a lot to the great co-authors! Esp
@rne.bsky.social & Yuanqi Du.
bit.ly/plaid-proteins
🧵
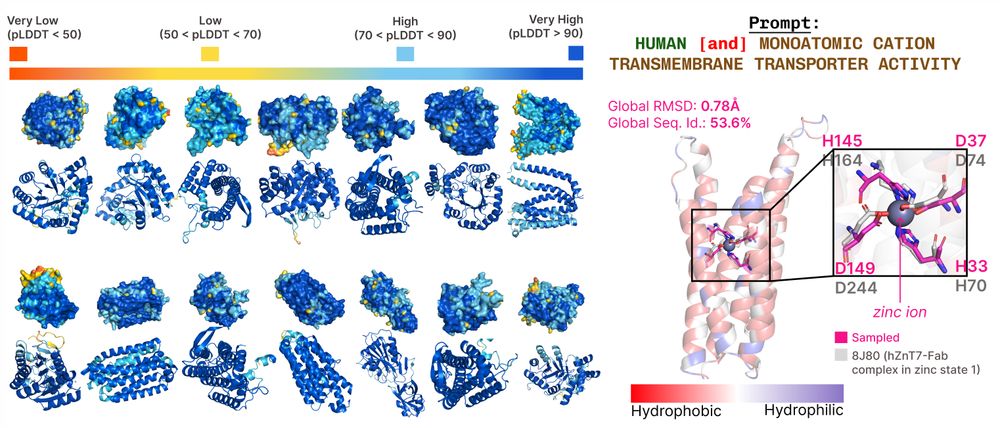
bit.ly/plaid-proteins
🧵
The call for talks is now open! Submit your abstract by January 5, 2024, at:
forms.gle/hu6BEWMN1BcR...

The call for talks is now open! Submit your abstract by January 5, 2024, at:
forms.gle/hu6BEWMN1BcR...
docs.bsky.app/docs/advance...
#AcademicSky #HigherEd #Altmetrics

docs.bsky.app/docs/advance...
#AcademicSky #HigherEd #Altmetrics