pubmed.ncbi.nlm.nih.gov/41379552/
introduces a new single-cell method named PICASSO: Phylogenetic Inference of CNA in Single-cell RNA [...]
github.com/dpeerlab/pic...

pubmed.ncbi.nlm.nih.gov/41379552/
introduces a new single-cell method named PICASSO: Phylogenetic Inference of CNA in Single-cell RNA [...]
github.com/dpeerlab/pic...

pubmed.ncbi.nlm.nih.gov/41379552/
introduces a new single-cell method named PICASSO: Phylogenetic Inference of CNA in Single-cell RNA [...]
github.com/dpeerlab/pic...


John Marioni, Head of Computation at Genentech Research and Early Development and former head of EBI
🧵
#scverse #scverse2025 #AI #DrugDevelopment #MachineLearning #ComputationalBiology #Biotech #Pharma #Genentech

John Marioni, Head of Computation at Genentech Research and Early Development and former head of EBI
🧵
#scverse #scverse2025 #AI #DrugDevelopment #MachineLearning #ComputationalBiology #Biotech #Pharma #Genentech
Details below
careersearch.stanford.edu/jobs/computa...
Plz RT


Details below
careersearch.stanford.edu/jobs/computa...
Plz RT
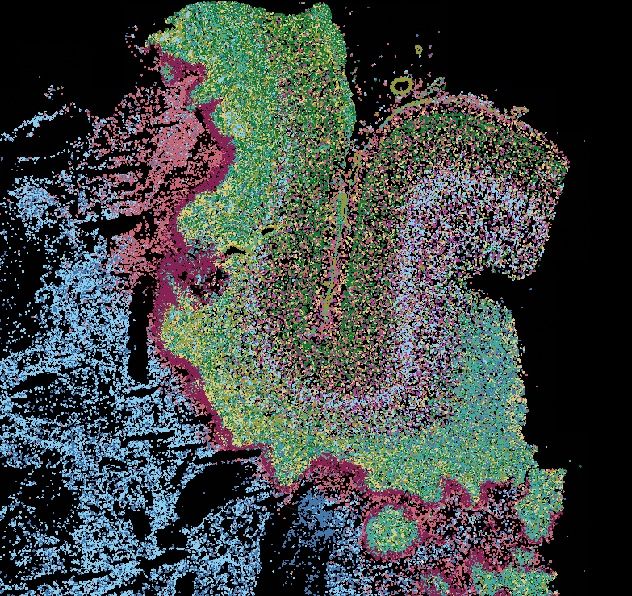
📖 Read: bit.ly/4mufVnZ @columbiaseas.bsky.social @columbiacancer.bsky.social @mskcancercenter.bsky.social
📖 Read: bit.ly/4mufVnZ @columbiaseas.bsky.social @columbiacancer.bsky.social @mskcancercenter.bsky.social
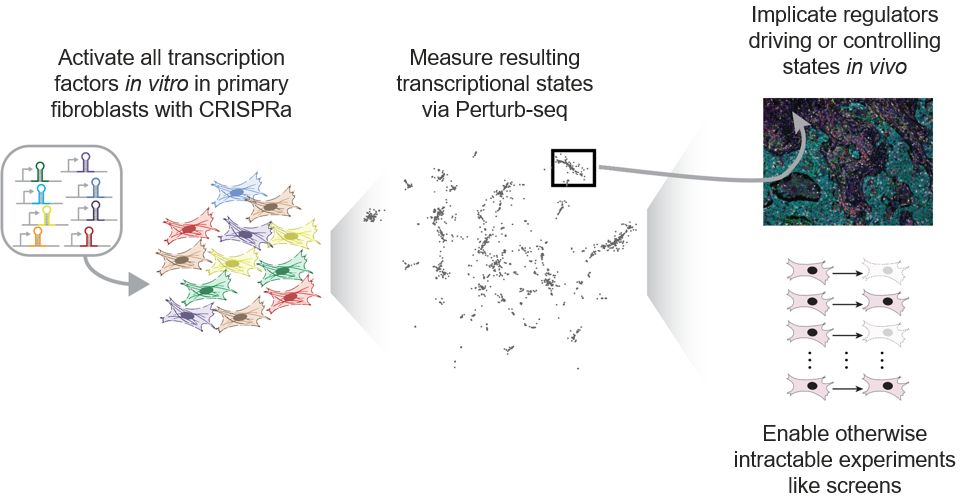
www.cell.com/cell-systems...
www.cell.com/cell-systems...


📅 July 22 | 🕑 15:50 | 📍 Room 01A
www.youtube.com/watch?v=8t8R...

www.youtube.com/watch?v=8t8R...

embl.wd103.myworkdayjobs.com/de-DE/EMBL/d...

grants.nih.gov/news-events/...

grants.nih.gov/news-events/...

@vanallenlab.bsky.social, who shared his work using AI to enhance precision cancer medicine.


DAMUTA has independent Damage and Misrepair signatures whose activities are more interpretable and more predictive of DNA repair defects, than COSMIC SBS signatures 🧬🖥️🧪
www.biorxiv.org/content/10.1...

DAMUTA has independent Damage and Misrepair signatures whose activities are more interpretable and more predictive of DNA repair defects, than COSMIC SBS signatures 🧬🖥️🧪
www.biorxiv.org/content/10.1...
@mskcancercenter.bsky.social is expanding and looking for ML-oriented computational biologists. Join our team based in NYC to work on cutting-edge single-cell & spatial transcriptomics methods in collaboration with leading research labs. bit.ly/4lTEr2t
@mskcancercenter.bsky.social is expanding and looking for ML-oriented computational biologists. Join our team based in NYC to work on cutting-edge single-cell & spatial transcriptomics methods in collaboration with leading research labs. bit.ly/4lTEr2t
youtu.be/8YhJM6zxYDw?...

youtu.be/8YhJM6zxYDw?...
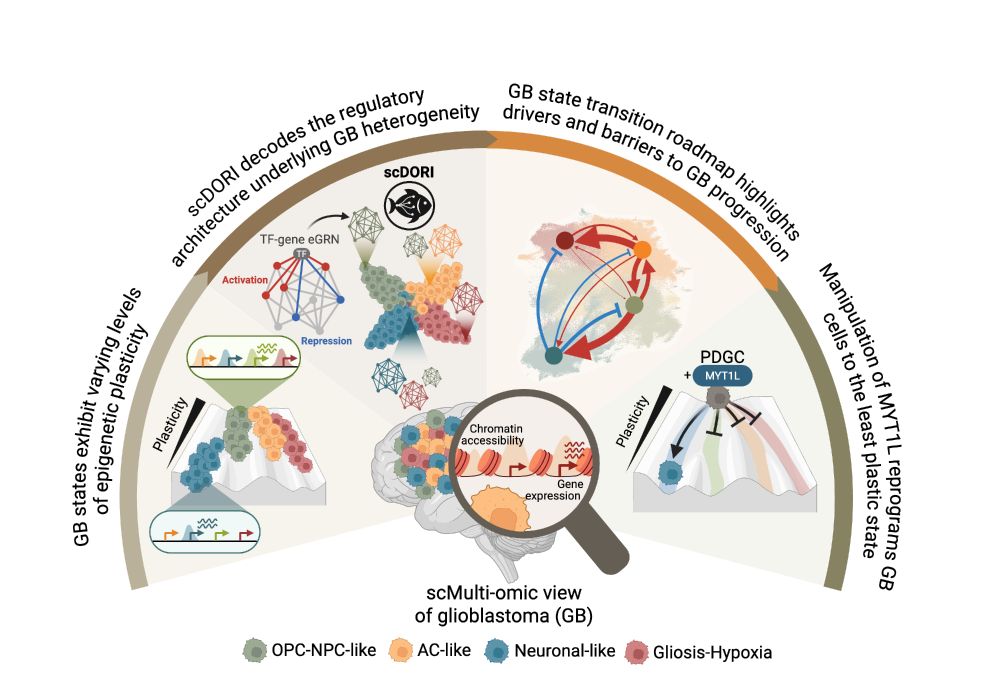
Our new method scDORI decodes plasticity regulators at single-cell multi-omic resolution — scaling to millions of cells and uncovering the regulatory logic driving glioblastoma heterogeneity.
🔍 Dive into the thread below and the preprint for all the insights!

Our new method scDORI decodes plasticity regulators at single-cell multi-omic resolution — scaling to millions of cells and uncovering the regulatory logic driving glioblastoma heterogeneity.
🔍 Dive into the thread below and the preprint for all the insights!