

For those who missed it, the introduction thread of REINDEER2
bsky.app/profile/npma...

For those who missed it, the introduction thread of REINDEER2
bsky.app/profile/npma...

doi.org/10.1038/s415...
@benjwoodcroft.bsky.social @rhysnewell.bsky.social
🧵1/6

doi.org/10.1038/s415...
@benjwoodcroft.bsky.social @rhysnewell.bsky.social
🧵1/6
I'm looking for studies where both Illumina and ONT sequencing were performed on the same samples from soil, human, ruminent, and other sample types for comparison. Bonus if those studies include PacBio data.
Please help and share!
I'm looking for studies where both Illumina and ONT sequencing were performed on the same samples from soil, human, ruminent, and other sample types for comparison. Bonus if those studies include PacBio data.
Please help and share!
academic.oup.com/bioinformati...

academic.oup.com/bioinformati...

www.nature.com/articles/d41...

www.nature.com/articles/d41...
👉 Preprint www.biorxiv.org/content/10.1...
👉 Github github.com/rolandfaure/...

👉 Preprint www.biorxiv.org/content/10.1...
👉 Github github.com/rolandfaure/...
Plasmid genomes were resolved using #PacBio HiFi sequencing with hifiasm-meta for #metagenome assembly. Host association was detected using epigenetic signals.
doi.org/10.1101/2025...

Plasmid genomes were resolved using #PacBio HiFi sequencing with hifiasm-meta for #metagenome assembly. Host association was detected using epigenetic signals.
doi.org/10.1101/2025...
Thread 1/n

Thread 1/n

Nanopore's getting accurate, but
1. Can this lead to better metagenome assemblies?
2. How, algorithmically, to leverage them?
with co-author Max Marin @mgmarin.bsky.social, supervised by Heng Li @lh3lh3.bsky.social
1 / N
Nanopore's getting accurate, but
1. Can this lead to better metagenome assemblies?
2. How, algorithmically, to leverage them?
with co-author Max Marin @mgmarin.bsky.social, supervised by Heng Li @lh3lh3.bsky.social
1 / N
metaMDBG (@gaetanbenoit.bsky.social) and Myloasm (@jimshaw.bsky.social) have had recent releases, so I updated the benchmarks from the Autocycler paper:
rrwick.github.io/2025/09/23/a...
Both tools improved considerably! Time to update your conda environments 😄
metaMDBG (@gaetanbenoit.bsky.social) and Myloasm (@jimshaw.bsky.social) have had recent releases, so I updated the benchmarks from the Autocycler paper:
rrwick.github.io/2025/09/23/a...
Both tools improved considerably! Time to update your conda environments 😄


Logan now democratizes efficient access to the world’s most comprehensive genetics dataset. Free and open.
doi.org/10.1101/2024...

Logan now democratizes efficient access to the world’s most comprehensive genetics dataset. Free and open.
doi.org/10.1101/2024...
- 30-40% potential reduction in memory with approximately the same runtime.
- Breaking changes to indexing and searching databases
Calculate ANI for contigs, genomes -- even search > 140k genomes. Pre-indexed GTDB-R226 available for download.

- 30-40% potential reduction in memory with approximately the same runtime.
- Breaking changes to indexing and searching databases
Calculate ANI for contigs, genomes -- even search > 140k genomes. Pre-indexed GTDB-R226 available for download.
github.com/lh3/longdust
github.com/lh3/longdust
www.medrxiv.org/content/10.1...
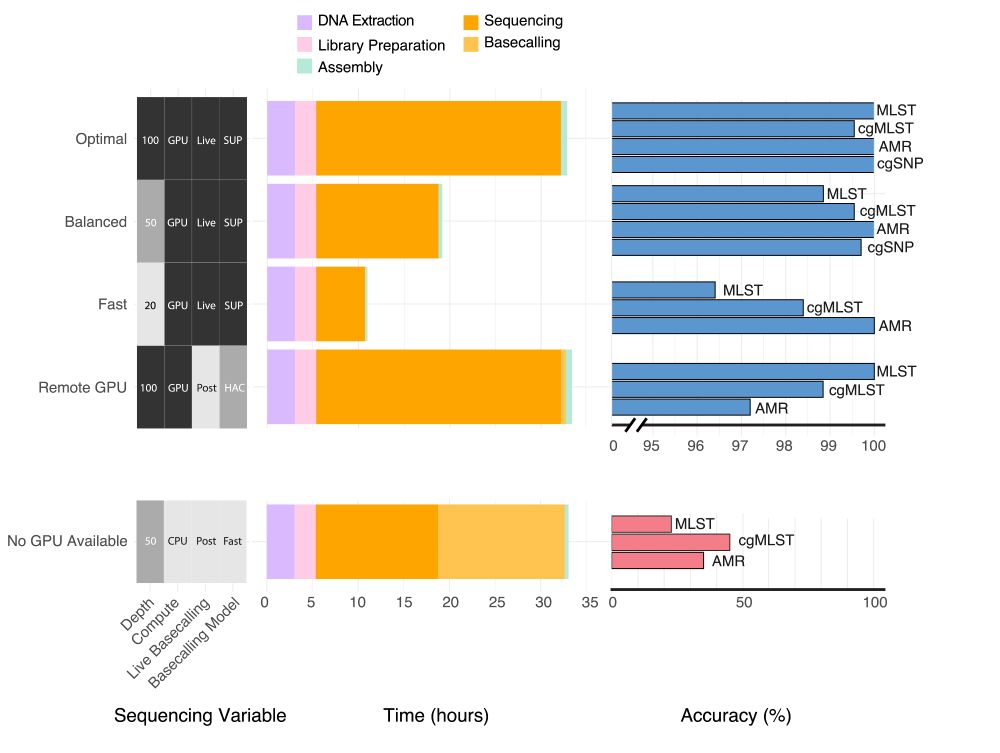
www.medrxiv.org/content/10.1...
@rayan.chiki.bsky.social
#Bioinformatics


@rayan.chiki.bsky.social
#Bioinformatics
- GTDB-R226 (143k prok. species)
- GlobDB-R226 (>300k prok. species, thanks @daanspeth.bsky.social )
- UHGV (Unified Human Gut Virome Catalog, thanks @apcamargo.bsky.social )
Must update sylph-tax; see docs (sylph-docs.github.io/sylph-tax/)
- GTDB-R226 (143k prok. species)
- GlobDB-R226 (>300k prok. species, thanks @daanspeth.bsky.social )
- UHGV (Unified Human Gut Virome Catalog, thanks @apcamargo.bsky.social )
Must update sylph-tax; see docs (sylph-docs.github.io/sylph-tax/)
Ever need to search for approximate matches of short DNA strings?
Sassy is the tool to use!
Available now wherever you get your code
With @rickbitloo.bsky.social
curiouscoding.nl/papers/sassy...
github.com/ragnarGrootK...
Ever need to search for approximate matches of short DNA strings?
Sassy is the tool to use!
Available now wherever you get your code
With @rickbitloo.bsky.social
curiouscoding.nl/papers/sassy...
github.com/ragnarGrootK...


