https://sites.google.com/view/guifengwei/
www.nature.com/articles/s41...

www.nature.com/articles/s41...
www.cell.com/molecular-ce...

www.cell.com/molecular-ce...
👉 We discover how sex and development signals are decoded at a single gene locus.
www.nature.com/articles/s41...
👇 Bluetorial

👉 We discover how sex and development signals are decoded at a single gene locus.
www.nature.com/articles/s41...
👇 Bluetorial

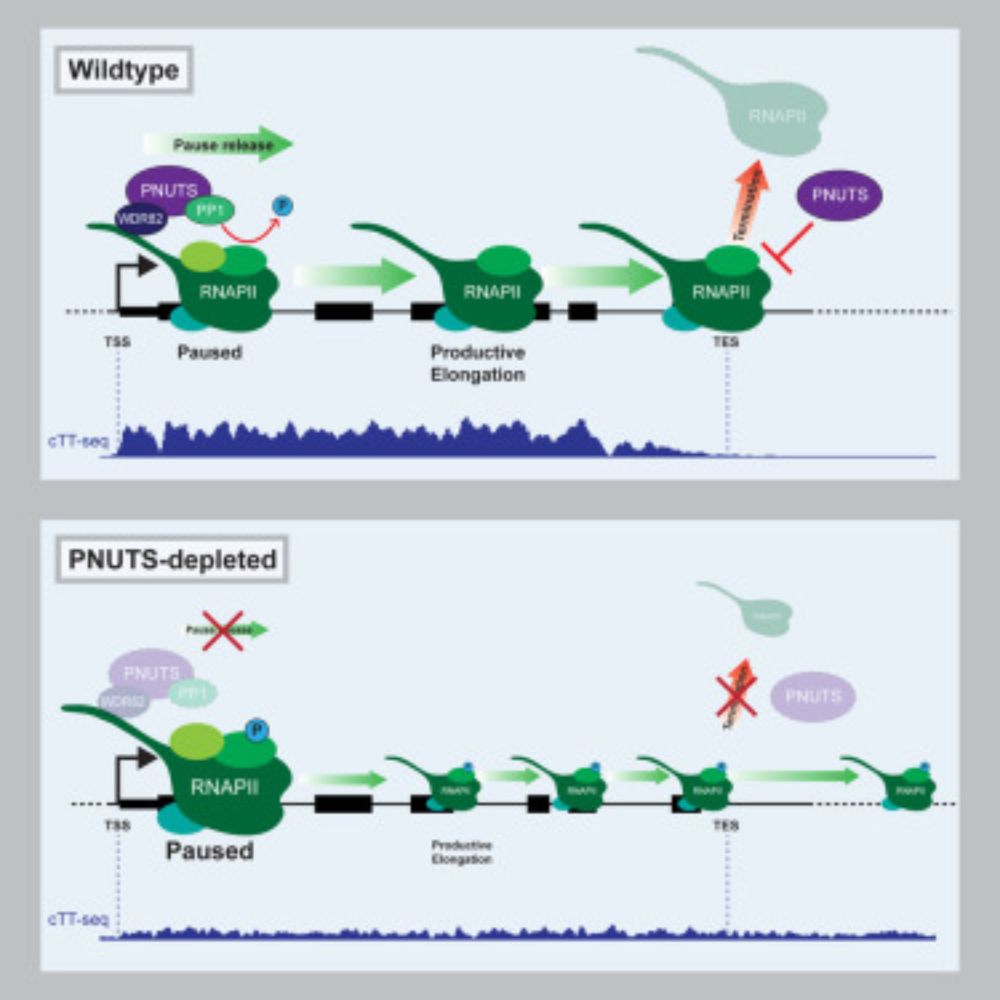
www.nature.com/articles/nme...



www.nature.com/articles/nme...
HCR-Proxy, a modular proximity labelling approach to profile local proteome composition around RNA at nanoscale, subcompartmental resolution. Thereby we resolved nested nucleolar layers and uncovered the grammar of spatial protein partitioning.
www.biorxiv.org/content/10.1...
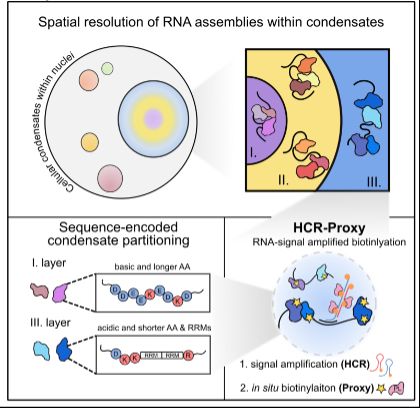
HCR-Proxy, a modular proximity labelling approach to profile local proteome composition around RNA at nanoscale, subcompartmental resolution. Thereby we resolved nested nucleolar layers and uncovered the grammar of spatial protein partitioning.
www.biorxiv.org/content/10.1...
She reveals how SMCHD1 dynamically engages chromatin, including the inactive X chromosome, to maintain gene silencing.
www.biorxiv.org/content/10.1...

She reveals how SMCHD1 dynamically engages chromatin, including the inactive X chromosome, to maintain gene silencing.
www.biorxiv.org/content/10.1...

www.biorxiv.org/content/10.1...
www.biorxiv.org/content/10.1...
We've lost a fantastic colleague, mentor & great friend to many. There was really no-one else like him. Working with him was so much fun & I owe him a lot. All my thoughts are with his family.
www.merton.ox.ac.uk/news/remembe...

blog.addgene.org/des...

blog.addgene.org/des...
www-nature-com.insb.bib.cnrs.fr/articles/s41...
It results from years of hard work by the team and long-time collaborators, with special thanks to PhD student Clara Roidor and collaborator Laurène Syx.

www-nature-com.insb.bib.cnrs.fr/articles/s41...
It results from years of hard work by the team and long-time collaborators, with special thanks to PhD student Clara Roidor and collaborator Laurène Syx.