Studying tandem repeats in single-cell contexts w/ @dgmacarthur.bsky.social
📄 www.medrxiv.org/content/10.1...

📄 www.medrxiv.org/content/10.1...
www.biorxiv.org/cgi/content/...

www.biorxiv.org/cgi/content/...

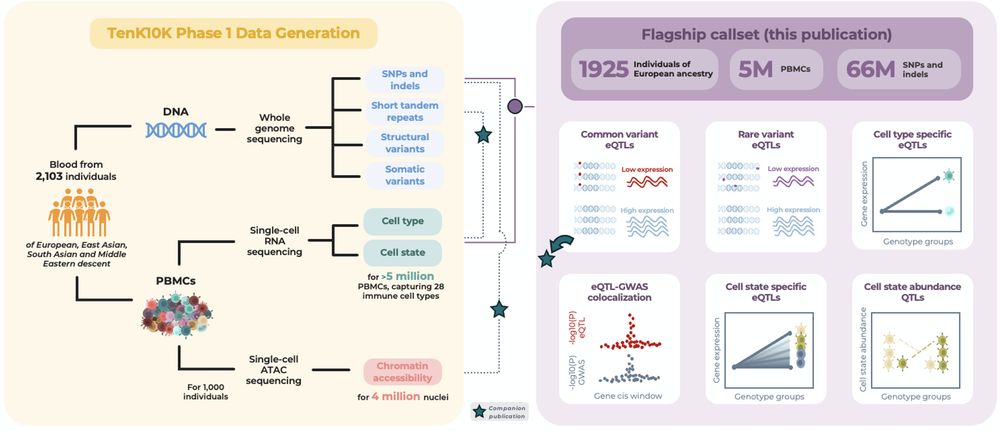
We explored causal effects of gene expression in immune cell types on complex traits and diseases by combining single-cell expression quantitative trait loci (sc-eQTL) mapping in 5M+ cells from 1,925 donors in TenK10K study and GWAS. 🧵

🧵👇 (1/n)

We describe the clinical phenotype of a recessive NDD associated with biallelic variants in RNU4-2 🧬
See 🧵👇
tinyurl.com/3j9r56s8
@rociorius.bsky.social @yuyangchen.bsky.social @gregfindlay.bsky.social @dgmacarthur.bsky.social @cassimons.bsky.social @nickywhiffin.bsky.social

We describe the clinical phenotype of a recessive NDD associated with biallelic variants in RNU4-2 🧬
See 🧵👇

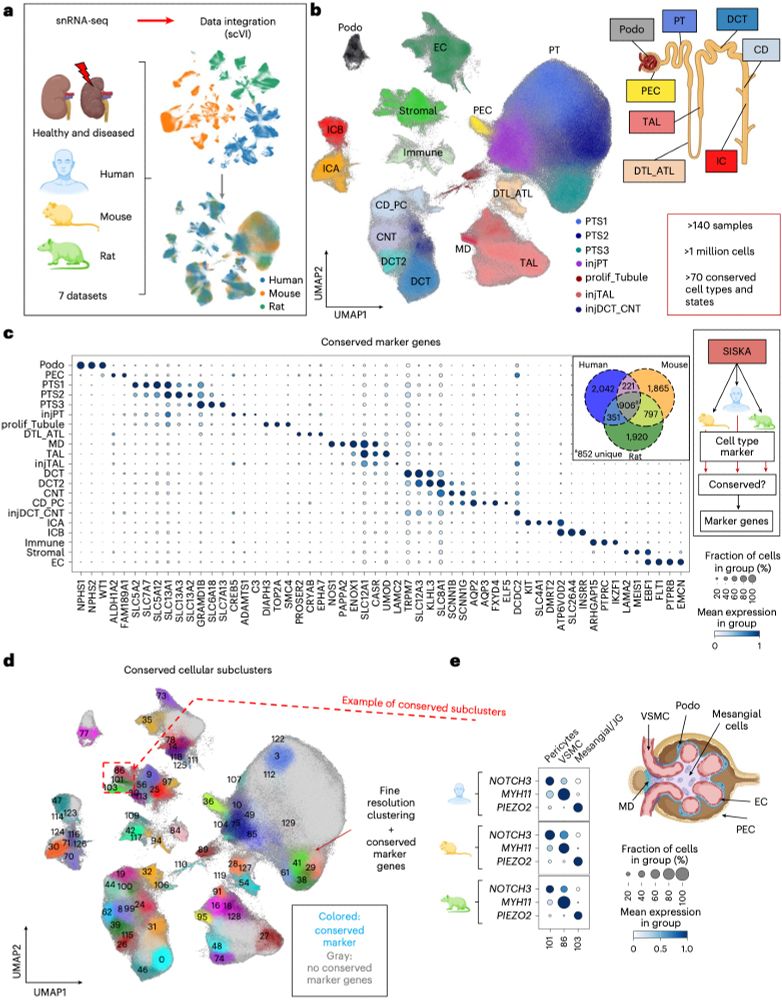
🚶♂️🐁🐀Cross-species 🫘kidney pathway dysregulation via #singlecell functional profiling of individual samples — opening new therapeutic avenues.
🔗 www.nature.com/articles/s41...
🧪 #NephSky #MedSky
🚶♂️🐁🐀Cross-species 🫘kidney pathway dysregulation via #singlecell functional profiling of individual samples — opening new therapeutic avenues.
🔗 www.nature.com/articles/s41...
🧪 #NephSky #MedSky

Our new paper by Mannion et al. takes a systematic look at "hidden enhancers" and why they remain so hard to find. With @mosterwalder.bsky.social, @jlopezrios.bsky.social & many more
www.nature.com/articles/s41...


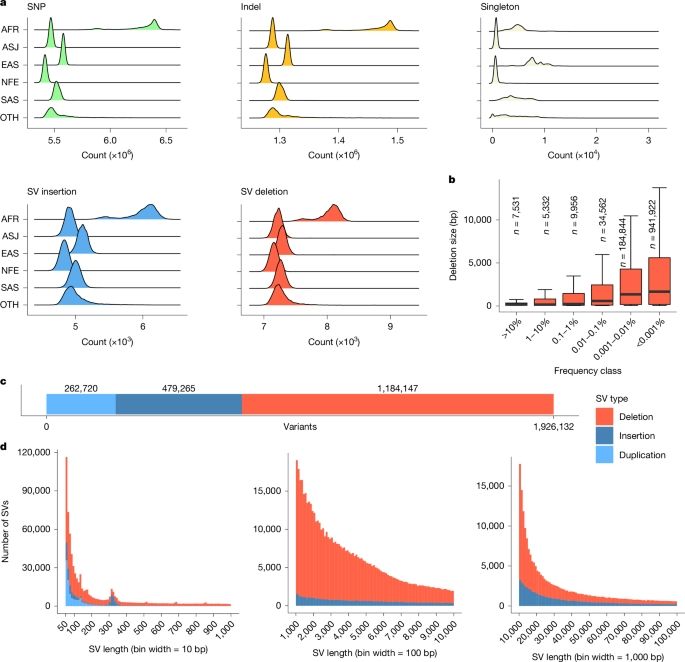




@gosiatrynka.bsky.social
@dgmacarthur.bsky.social
@bpasaniuc.bsky.social
@tuuliel.bsky.social
@hilarycmartin.bsky.social
@sashagusevposts.bsky.social
@zkutalik.bsky.social
@mashaals.bsky.social
@alemedinarivera.bsky.social

@gosiatrynka.bsky.social
@dgmacarthur.bsky.social
@bpasaniuc.bsky.social
@tuuliel.bsky.social
@hilarycmartin.bsky.social
@sashagusevposts.bsky.social
@zkutalik.bsky.social
@mashaals.bsky.social
@alemedinarivera.bsky.social
medrxiv.org/content/10.1101/2025.06.24.25330216
The main output from my PhD is finally public and we’re SUPER excited about the findings! If you’re interested in what we learnt about IBD with a massive 700+ sample sc-eQTL dataset of the gut, read on!

medrxiv.org/content/10.1101/2025.06.24.25330216
The main output from my PhD is finally public and we’re SUPER excited about the findings! If you’re interested in what we learnt about IBD with a massive 700+ sample sc-eQTL dataset of the gut, read on!
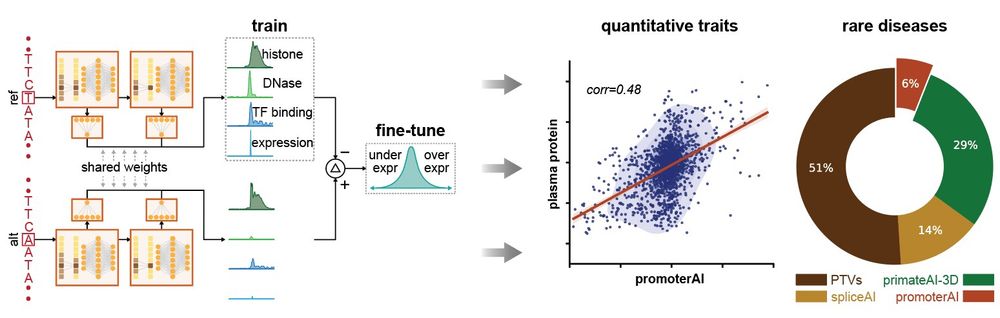
www.biorxiv.org/content/10.1...

www.biorxiv.org/content/10.1...
github.com/populationge...
A superb collaboration with @dgmacarthur.bsky.social @cassimons.bsky.social @heidirehm.bsky.social @ksamocha.bsky.social and many more!

github.com/populationge...
A superb collaboration with @dgmacarthur.bsky.social @cassimons.bsky.social @heidirehm.bsky.social @ksamocha.bsky.social and many more!

I’ll be sharing some findings on tandem repeats #TRs in single cell contexts using TenK10K Phase 1 on Thursday at 11:55.
I’ll be sharing some findings on tandem repeats #TRs in single cell contexts using TenK10K Phase 1 on Thursday at 11:55.
Details 👉 petercallen.github.io/2025-OSC-hac...
#OzSingleCell #ozsinglecell25 @ozsinglecell.bsky.social

Details 👉 petercallen.github.io/2025-OSC-hac...
#OzSingleCell #ozsinglecell25 @ozsinglecell.bsky.social
Join me, @dgmacarthur.bsky.social, and the CPG Rare Disease Program:
career10.successfactors.com/career?caree...
Join me, @dgmacarthur.bsky.social, and the CPG Rare Disease Program:
career10.successfactors.com/career?caree...

🧵👇 (1/n)