

Apply by Feb 24: email [email protected] with “[Open-25]” in the subject. DM for details!
lnkd.in/eD-ueQmV
lnkd.in/eASJEZAW

lnkd.in/eD-ueQmV
lnkd.in/eASJEZAW
⏳Application deadline extended until March 14th!
👇 Open positions & application info:
🧬 Passionate about #SyntheticBiology? Based in #Europe? Help grow our community!
We are opening new positions, check them out & apply here 👉 linkedin.com/feed/update/...
#SynBio #ScienceCommunication #Policy

⏳Application deadline extended until March 14th!
👇 Open positions & application info:
ProtComposer: Compositional Protein Structure Generation with 3D Ellipsoids arxiv.org/abs/2503.05025
Condition on your 3D layout (of ellipsoids) to generate proteins like this or to get better designability/diversity/novelty tradeoffs.
1/6
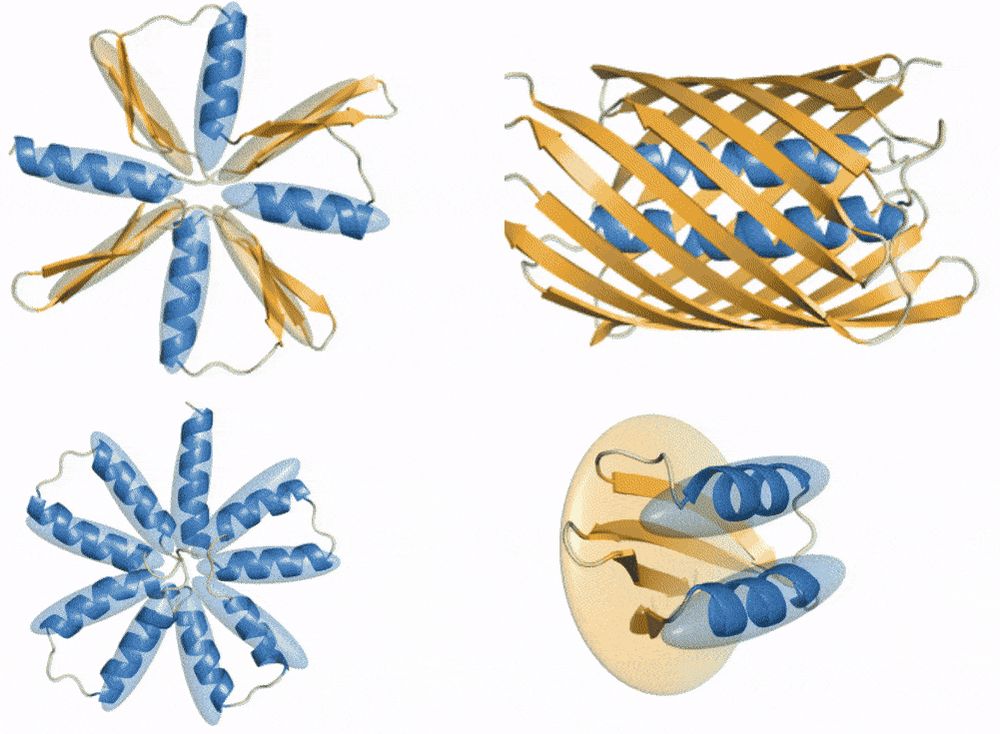
ProtComposer: Compositional Protein Structure Generation with 3D Ellipsoids arxiv.org/abs/2503.05025
Condition on your 3D layout (of ellipsoids) to generate proteins like this or to get better designability/diversity/novelty tradeoffs.
1/6
thisgenomiclife.substack.com/p/can-genera...

thisgenomiclife.substack.com/p/can-genera...
Apply by Feb 24: email [email protected] with “[Open-25]” in the subject. DM for details!

rdcu.be/d9H92
Happy to share a conceptual schematic for a “phrinter (phage printer)” 😊
Many thanks to all co-authors, editors and reviewers!

rdcu.be/d9H92
Happy to share a conceptual schematic for a “phrinter (phage printer)” 😊
Many thanks to all co-authors, editors and reviewers!
Attend the premier conference at the intersection of ML & Bio, share your research and make lasting connections!
Submission deadline: June 1
More details: mlcb.github.io
Help spread the word—please RT! #MLCB2025
Attend the premier conference at the intersection of ML & Bio, share your research and make lasting connections!
Submission deadline: June 1
More details: mlcb.github.io
Help spread the word—please RT! #MLCB2025
Apply by Feb 24: email [email protected] with “[Open-25]” in the subject. DM for details!

Apply by Feb 24: email [email protected] with “[Open-25]” in the subject. DM for details!
@itamarchinn.bsky.social @pgmikhael.bsky.social
www.science.org/doi/10.1126/...



@itamarchinn.bsky.social @pgmikhael.bsky.social
www.science.org/doi/10.1126/...
In new work led by @liambai.bsky.social and me, we explore how sparse autoencoders can help us understand biology—going from mechanistic interpretability to mechanistic biology.
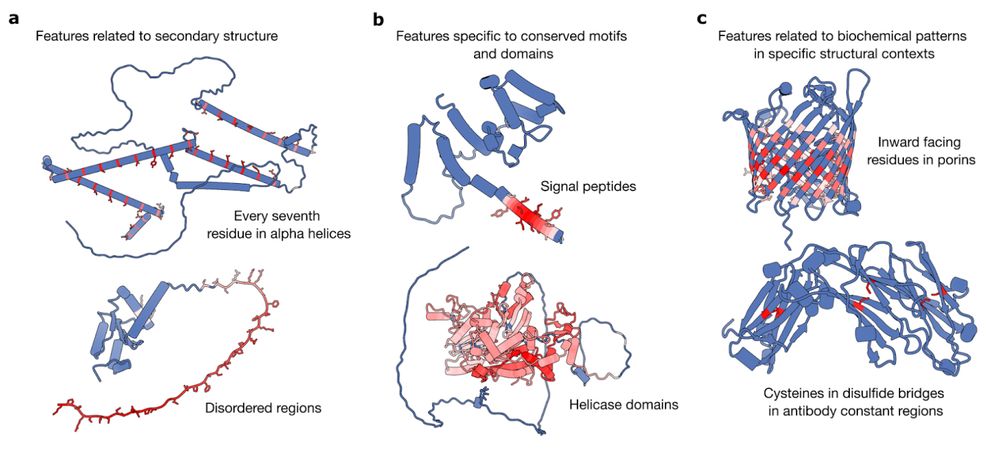
www.biorxiv.org/content/10.1...
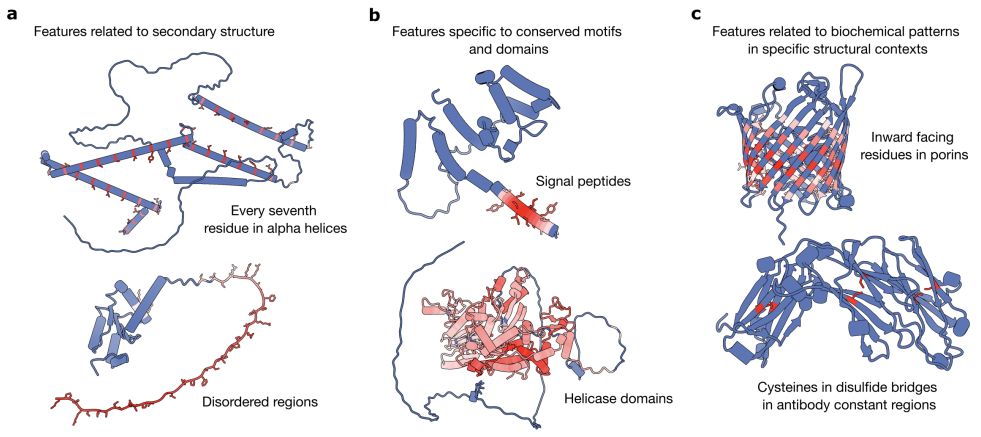
www.biorxiv.org/content/10.1...
25/n
www.science.org/doi/10.1126/...

www.science.org/doi/10.1126/...


