

Huge thanks to @danielibrahim.bsky.social, @arnaudkr.bsky.social & @stemundi.bsky.social and fantastic people in their labs — what a journey! 🧪🔬
We very happy to share our latest work trying to understand enhancer-promoter compatibility.
I am very excited about the results of @blanka-majchrzycka.bsky.social, which changed the way I think about promoters
www.biorxiv.org/content/10.1...

Huge thanks to @danielibrahim.bsky.social, @arnaudkr.bsky.social & @stemundi.bsky.social and fantastic people in their labs — what a journey! 🧪🔬
www.biorxiv.org/content/10.1...
A🧵👇
www.biorxiv.org/content/10.1...
A🧵👇
The main story of my PhD deals with exactly that question, and is now published in Science! ✨
www.science.org/doi/10.1126/...
My amazing co-author and friend @mathiaseder.bsky.social summarized the highlights for you
The main story of my PhD deals with exactly that question, and is now published in Science! ✨
www.science.org/doi/10.1126/...
My amazing co-author and friend @mathiaseder.bsky.social summarized the highlights for you

register here for the entire series: us06web.zoom.us/webinar/regi...

register here for the entire series: us06web.zoom.us/webinar/regi...
"De-novo promoters emerge more readily from random DNA than from genomic DNA"
This project is the accumulation of 4 years of work, and lays the foundation for my future group. In short, we… (1/4)

"De-novo promoters emerge more readily from random DNA than from genomic DNA"
This project is the accumulation of 4 years of work, and lays the foundation for my future group. In short, we… (1/4)
www.nature.com/articles/s41...

www.nature.com/articles/s41...
We are thrilled to announce that our lab’s first preprint is out!
”Whole-genome single-cell multimodal history tracing to reveal cell identity transition”
We report HisTrac-seq, a multiomic single-cell molecular recording platform.
www.biorxiv.org/content/10.1...
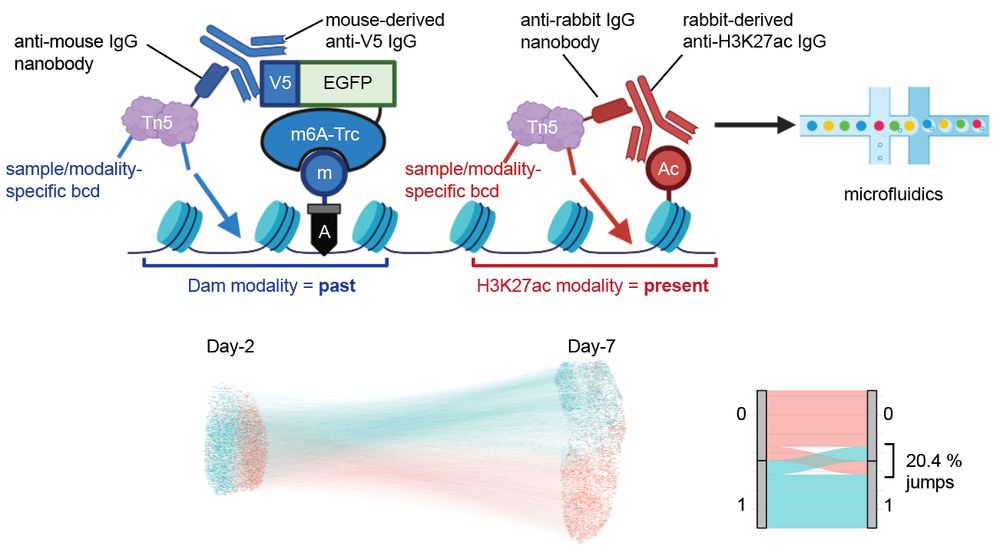
We are thrilled to announce that our lab’s first preprint is out!
”Whole-genome single-cell multimodal history tracing to reveal cell identity transition”
We report HisTrac-seq, a multiomic single-cell molecular recording platform.
www.biorxiv.org/content/10.1...



In case you haven't joined any of the previous 2025 sessions, here is registration link:
us06web.zoom.us/webinar/regi...

In case you haven't joined any of the previous 2025 sessions, here is registration link:
us06web.zoom.us/webinar/regi...
Check out our paper - fresh off the press!!!
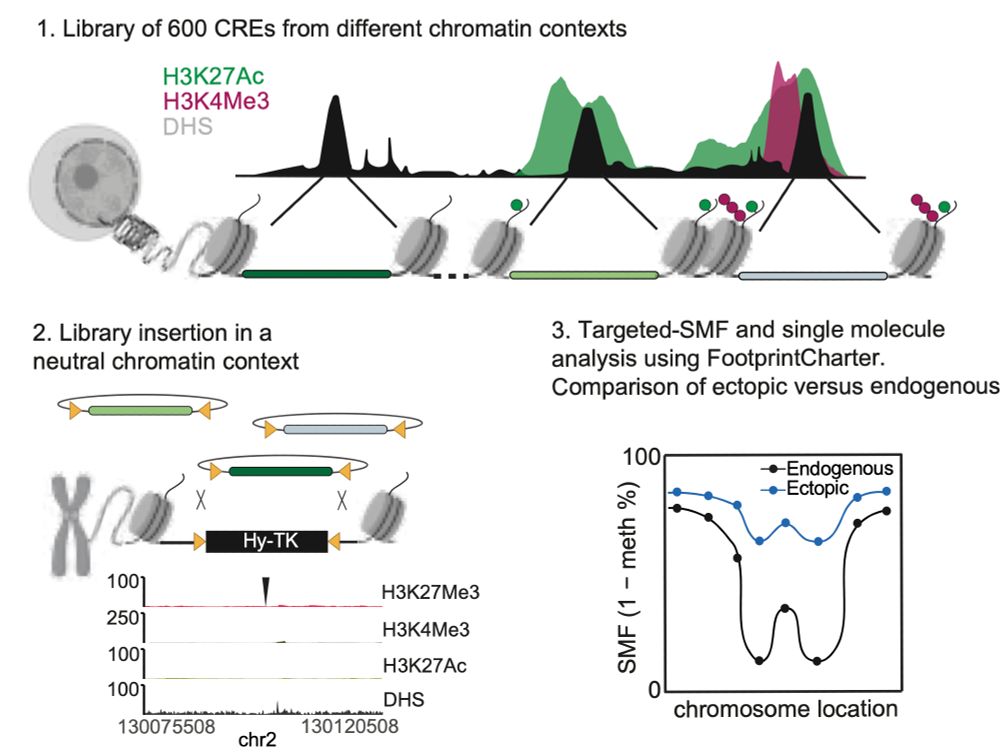
We find widespread functional conservation of enhancers in absence of sequence homology
Including: a bioinformatic tool to map sequence-diverged enhancers!
rdcu.be/enVDN
github.com/tobiaszehnde...

Check out our paper - fresh off the press!!!
We find widespread functional conservation of enhancers in absence of sequence homology
Including: a bioinformatic tool to map sequence-diverged enhancers!
rdcu.be/enVDN
github.com/tobiaszehnde...
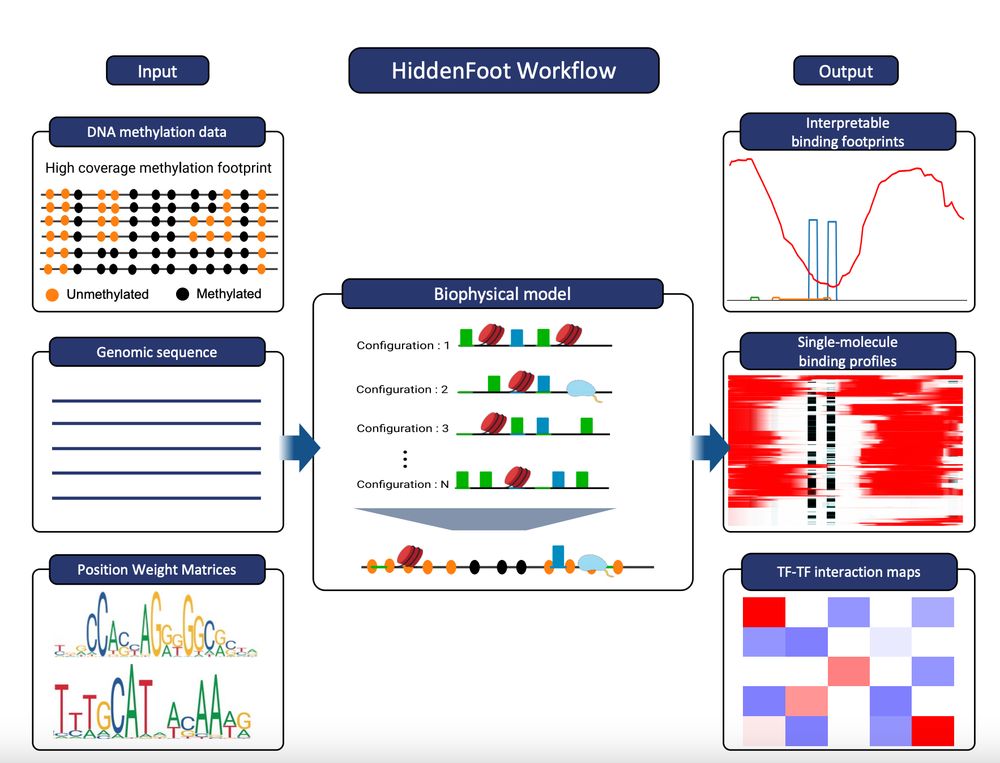
You can register at: us06web.zoom.us/webinar/regi...
You can register at: us06web.zoom.us/webinar/regi...
tissue-specific huntingtin interactomes, gene product diversity evolution
Cover: mammalian promoters characterised by low RNA pol II occupancy and high turnover @kasitc.bsky.social @molinalab.bsky.social @arnaudkr.bsky.social

tissue-specific huntingtin interactomes, gene product diversity evolution
Cover: mammalian promoters characterised by low RNA pol II occupancy and high turnover @kasitc.bsky.social @molinalab.bsky.social @arnaudkr.bsky.social
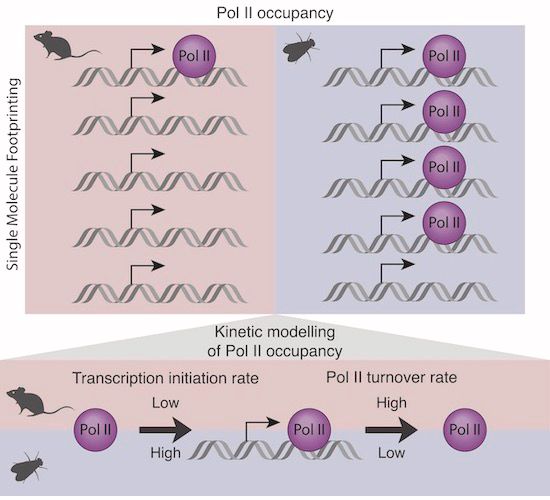


Using single-molecule footprinting, we showed that Pol II occupancy is lower at mouse than fly promoters. This low Pol II occupancy is explained by high Pol II turnover and low transcription initiation at mouse promoters.
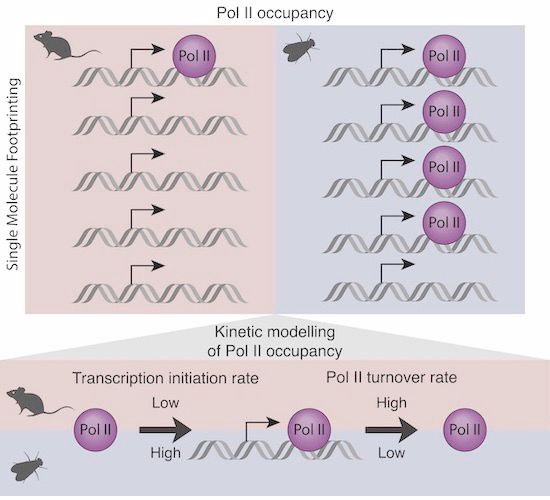
Using single-molecule footprinting, we showed that Pol II occupancy is lower at mouse than fly promoters. This low Pol II occupancy is explained by high Pol II turnover and low transcription initiation at mouse promoters.

