
@zymoresearch.bsky.social mocks and metagenomes ( github.com/Kirk3gaard/M...). Basecalled reads with methylation is in @enasequence.bsky.social . Eventually the pod5s might go there as well.
Happy Friday everyone.

Happy Friday everyone.
apnews.com/article/mode...

apnews.com/article/mode...
me: did you reverse complement the genome sequence?
claude: i'm a dumbass
me: did you reverse complement the genome sequence?
claude: i'm a dumbass

www.nature.com/articles/s41...

www.nature.com/articles/s41...






The E-STEEM programme call opens January 7th, 2026 and closes March 3rd, 2026.
careers.univie.ac.at/en/postdoc/e...
I participate as a host...
The E-STEEM programme call opens January 7th, 2026 and closes March 3rd, 2026.
careers.univie.ac.at/en/postdoc/e...
I participate as a host...



I'm looking for studies where both Illumina and ONT sequencing were performed on the same samples from soil, human, ruminent, and other sample types for comparison. Bonus if those studies include PacBio data.
Please help and share!
I'm looking for studies where both Illumina and ONT sequencing were performed on the same samples from soil, human, ruminent, and other sample types for comparison. Bonus if those studies include PacBio data.
Please help and share!
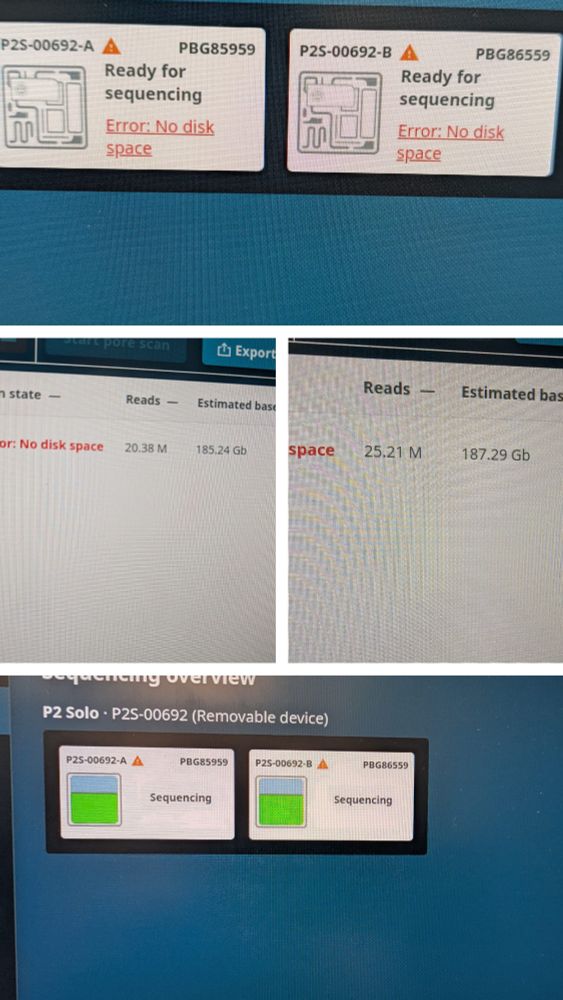
Let's find out! I created a form where you can register your yields so we can get a better overview. github.com/Kirk3gaard/O...
Let's find out! I created a form where you can register your yields so we can get a better overview. github.com/Kirk3gaard/O...


jobs.awi.de/Vacancies/20...
This is quite an exciting opportunity to push the boundaries of what is known regarding the molecular basis of the formation and demise of photosymbiotic relationships in marine habitats.
jobs.awi.de/Vacancies/20...
This is quite an exciting opportunity to push the boundaries of what is known regarding the molecular basis of the formation and demise of photosymbiotic relationships in marine habitats.


