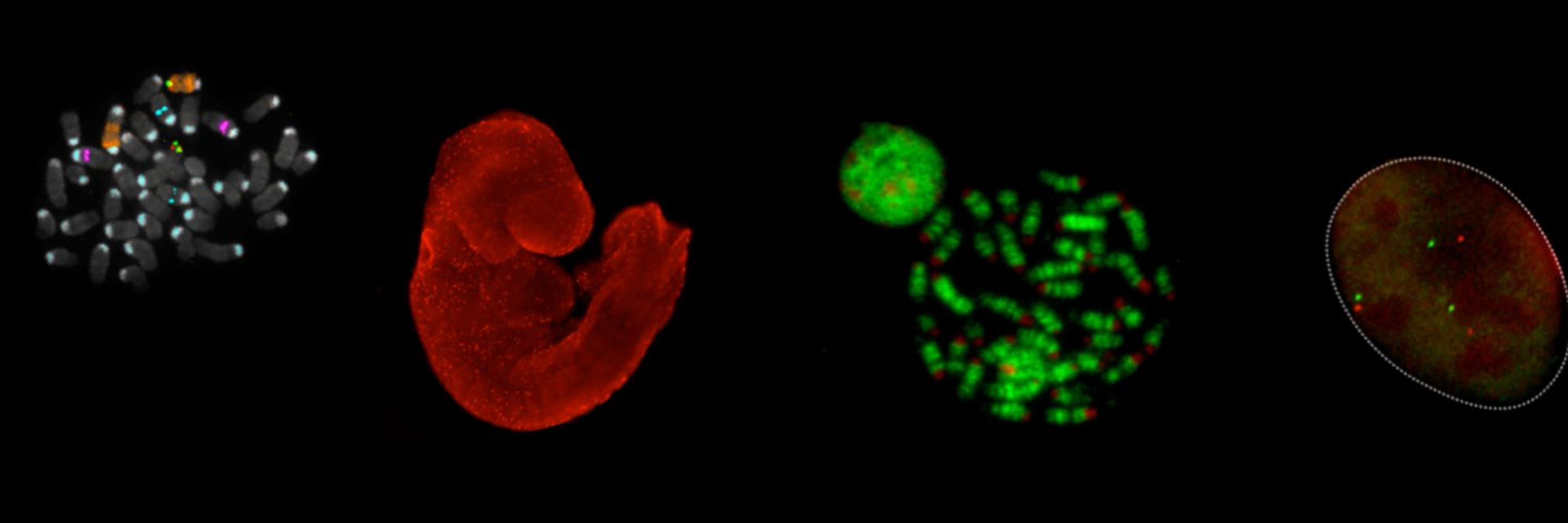
#gene_regulation #nuclear_organization #development | handkerchief user | views are mine alone, probably for the best
rochalab.nichd.nih.gov
tinyurl.com/4rz8kusj
Yajie Zhu (PhD graduate) and Mariano Barbieri (postdoc, @spp2202.bsky.social Accelerator award) produced a package that will classify loops in your #3Dgenomics data based on (whichever) epigenomic data you have available. Keep reading...
1/n
www.biorxiv.org/content/10.6...
Yajie Zhu (PhD graduate) and Mariano Barbieri (postdoc, @spp2202.bsky.social Accelerator award) produced a package that will classify loops in your #3Dgenomics data based on (whichever) epigenomic data you have available. Keep reading...
1/n
www.biorxiv.org/content/10.6...
www.biorxiv.org/content/10.6...
www.biorxiv.org/content/10.6...
Led by @raquelrouco.bsky.social, this study establishes a new framework to study how enhancer landscapes act sequentially at developmental loci and are silenced to shape gene expression patterns. (1/n)
Led by @raquelrouco.bsky.social, this study establishes a new framework to study how enhancer landscapes act sequentially at developmental loci and are silenced to shape gene expression patterns. (1/n)

A histone ubiquitylation-based regulatory hub links stress/environmental signaling to heterochromatin self-propagation and epigenetic inheritance-reshaping how we think about development, drug resistance, and cancer
👉 nature.com/articles/s41586-025-09899-8

A histone ubiquitylation-based regulatory hub links stress/environmental signaling to heterochromatin self-propagation and epigenetic inheritance-reshaping how we think about development, drug resistance, and cancer
👉 nature.com/articles/s41586-025-09899-8

Our department @ucl-cdb.bsky.social is looking to support scientists seeking to establish their lab with a fellowship. Deadline: 30 Jan. Apply here: www.ucl.ac.uk/work-at-ucl/...
Our department @ucl-cdb.bsky.social is looking to support scientists seeking to establish their lab with a fellowship. Deadline: 30 Jan. Apply here: www.ucl.ac.uk/work-at-ucl/...
Passionate about gene regulation, chromatin, and developmental biology? Just contact @radaiglesiaslab.bsky.social at @ibbtec.bsky.social 🧬✨
#PhD #3DGenome

Passionate about gene regulation, chromatin, and developmental biology? Just contact @radaiglesiaslab.bsky.social at @ibbtec.bsky.social 🧬✨
#PhD #3DGenome
We now offer a 4-years PhD contract to investigate whether the dirsuption of this regulatory mechanism can lead to congenital defects. More details 👇

We now offer a 4-years PhD contract to investigate whether the dirsuption of this regulatory mechanism can lead to congenital defects. More details 👇
doi.org/10.1016/j.ce...
We developed a new method called MCC ultra, which allows 3D chromatin structure to be visualised with a 1 base pair pixel size.

doi.org/10.1016/j.ce...
We developed a new method called MCC ultra, which allows 3D chromatin structure to be visualised with a 1 base pair pixel size.
Perfect timing for our paper from the lab of @toddmacfarlan.bsky.social to be out @natcomms.nature.com!!
…and I’m currently on the job market looking for a new scientific home!
www.nature.com/articles/s41...

Perfect timing for our paper from the lab of @toddmacfarlan.bsky.social to be out @natcomms.nature.com!!
…and I’m currently on the job market looking for a new scientific home!
www.nature.com/articles/s41...

💼 Un poste susceptible d’être vacant de Professeur en stabilité des génomes devrait s’ouvrir pour la rentrée 2026 au sein de l’Institut Jacques Monod
➡️ Informations concernant ce poste et la procédure de candidature 🔗 www.ijm.fr/poste-de-pro...



Was co-submitted with @allanaschooley.bsky.social @jobdekker.bsky.social whose paper should also come out soon
Brief thread 👇
Was co-submitted with @allanaschooley.bsky.social @jobdekker.bsky.social whose paper should also come out soon
Brief thread 👇
We very happy to share our latest work trying to understand enhancer-promoter compatibility.
I am very excited about the results of @blanka-majchrzycka.bsky.social, which changed the way I think about promoters
www.biorxiv.org/content/10.1...

We very happy to share our latest work trying to understand enhancer-promoter compatibility.
I am very excited about the results of @blanka-majchrzycka.bsky.social, which changed the way I think about promoters
www.biorxiv.org/content/10.1...
Check our latest preprint, led by @mmasoura.bsky.social, to find out!
www.biorxiv.org/content/10.1...

Check our latest preprint, led by @mmasoura.bsky.social, to find out!
www.biorxiv.org/content/10.1...
#IMPRS #IMPRSBAC @freieuniversitaet.bsky.social @maxplanck.de
Check out our projects and apply by Jan 7th, 2026!
www.molgen.mpg.de/IMPRSPhDproj...

In a new preprint, we share some clues about when, how, and why this happens!
www.biorxiv.org/content/10.1...

In a new preprint, we share some clues about when, how, and why this happens!
www.biorxiv.org/content/10.1...
www.biorxiv.org/content/10.1...

www.biorxiv.org/content/10.1...
Check out this preprint combining experiments and modelling by our colleagues and partners in crime @ucl-cdb.bsky.social:
www.biorxiv.org/content/10.1...

Check out this preprint combining experiments and modelling by our colleagues and partners in crime @ucl-cdb.bsky.social:
www.biorxiv.org/content/10.1...
Job posting: www.training.nih.gov/jobs/pdf-gb-...
Lab Website: www.niddk.nih.gov/research-fun...
NIH Trainee info: www.training.nih.gov

Job posting: www.training.nih.gov/jobs/pdf-gb-...
Lab Website: www.niddk.nih.gov/research-fun...
NIH Trainee info: www.training.nih.gov
www.biorxiv.org/content/10.1...
A🧵👇
www.biorxiv.org/content/10.1...
A🧵👇
www.biorxiv.org/content/10.1...
A tour-de-force by 1st authors Bruna Eckhardt & @palindromephd.bsky.social, focusing on chromatin replication. RTs welcome; tweetorial in 3,2...(1/n)

www.biorxiv.org/content/10.1...
A tour-de-force by 1st authors Bruna Eckhardt & @palindromephd.bsky.social, focusing on chromatin replication. RTs welcome; tweetorial in 3,2...(1/n)

