Learn more: www.biorxiv.org/content/10.1...

Learn more: www.biorxiv.org/content/10.1...



Time for a science thread! #TeamMassSpec #SpatialProteomics #AtheroSky 🧵
www.biorxiv.org/content/10.1...
Time for a science thread! #TeamMassSpec #SpatialProteomics #AtheroSky 🧵
www.biorxiv.org/content/10.1...
We used proteomics on Paranthropus robustus fossils from South Africa (~2 million years old) to get insights into biological sex and variation.
A great example of how LC-MS/MS can uncover new transformative info👇
Our paper on enamel proteins from Paranthropus robustus has finally been peer reviewed, please have a read here: www.science.org/doi/10.1126/...
Paranthropus robustus has been puzzling scientists since its discovery in 1938 in South Africa, where a high number of fossils have been found.
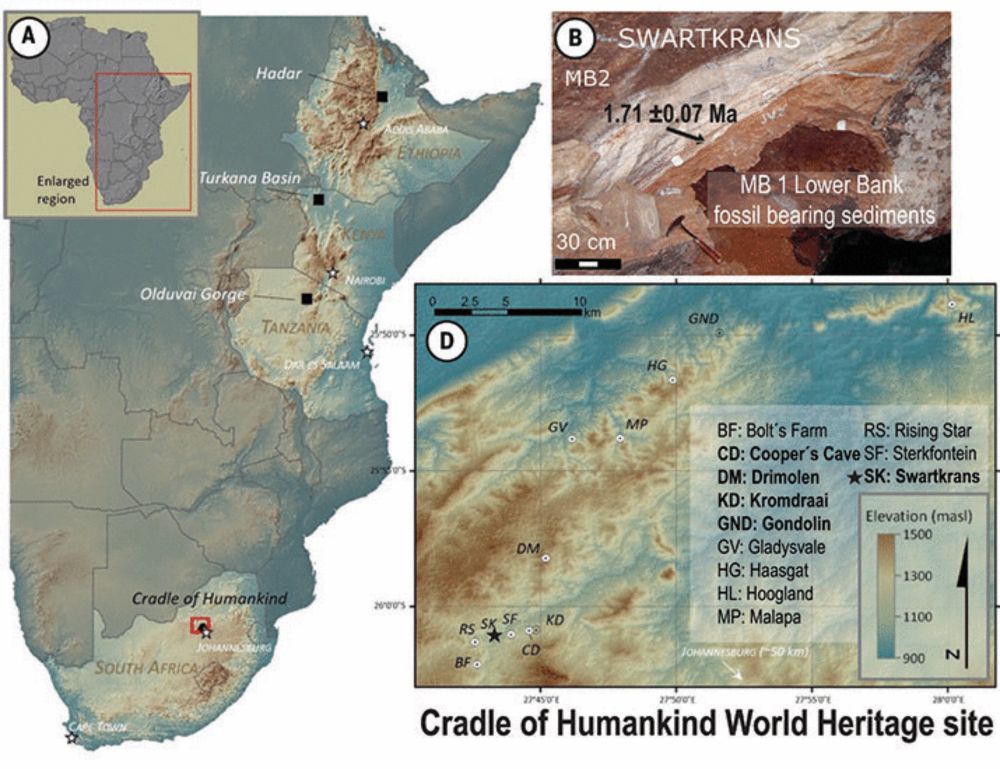
We used proteomics on Paranthropus robustus fossils from South Africa (~2 million years old) to get insights into biological sex and variation.
A great example of how LC-MS/MS can uncover new transformative info👇
---
#proteomics #prot-preprint

We've developed SC-pSILAC to simultaneously measure protein turnover and abundance in single cells, unlocking the first large-scale, 2D proteomic insights at single-cell resolution!
www.cell.com/cell/fulltex...
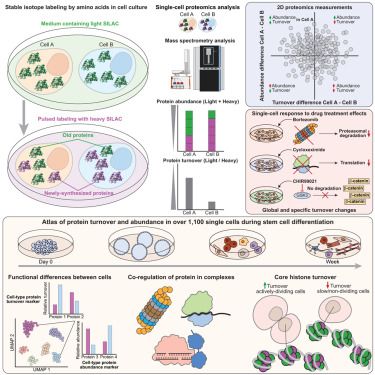
We've developed SC-pSILAC to simultaneously measure protein turnover and abundance in single cells, unlocking the first large-scale, 2D proteomic insights at single-cell resolution!
www.cell.com/cell/fulltex...
Read full study here: www.nature.com/articles/s41...




Read full study here: www.nature.com/articles/s41...
They then combined it with PhiSDM spectral processing, mitigating the resolution sacrifice.
www.biorxiv.org/content/10.1...
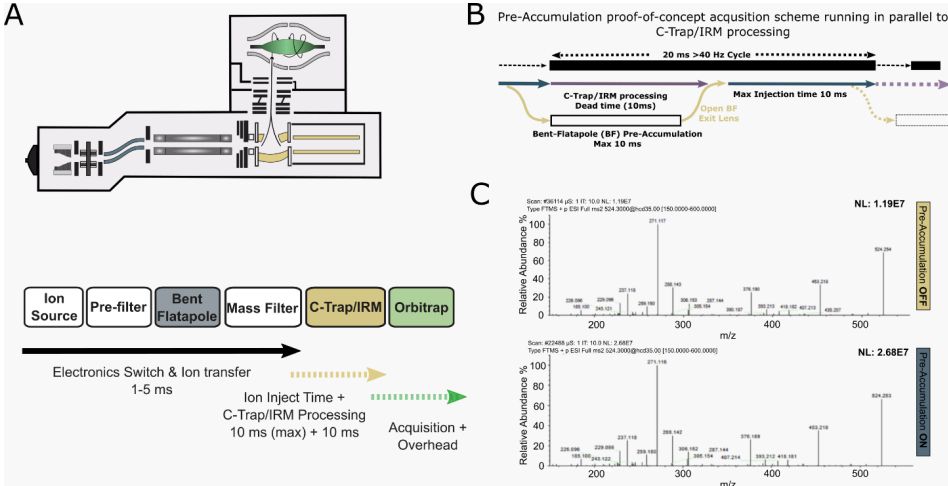
They then combined it with PhiSDM spectral processing, mitigating the resolution sacrifice.
www.biorxiv.org/content/10.1...
Protein degradation is a HUGE factor.
It accounts for up to 50 % of protein variation across proteins & tissue types.
www.biorxiv.org/content/10.1...
🧵
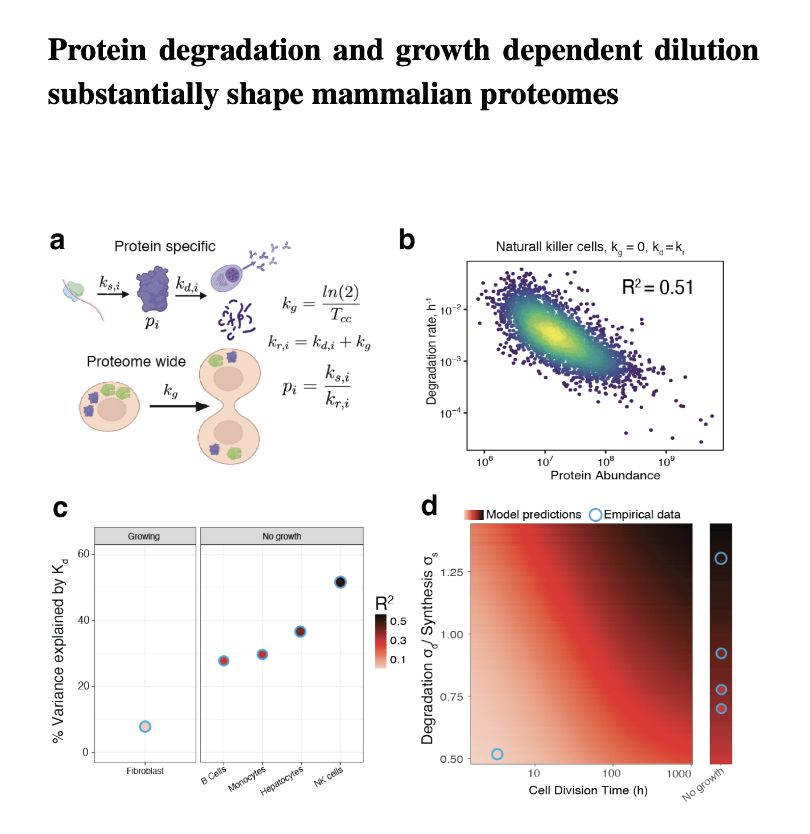
Protein degradation is a HUGE factor.
It accounts for up to 50 % of protein variation across proteins & tissue types.
www.biorxiv.org/content/10.1...
🧵
We evaluated how formaldehyde-based fixation preserve proteome state, drug response and cell integrity. Ultimately, cell fixation can facilitate access to #singlecell by enabeling sample shiping and prolonged sorting 📦
pubs.acs.org/doi/10.1021/...
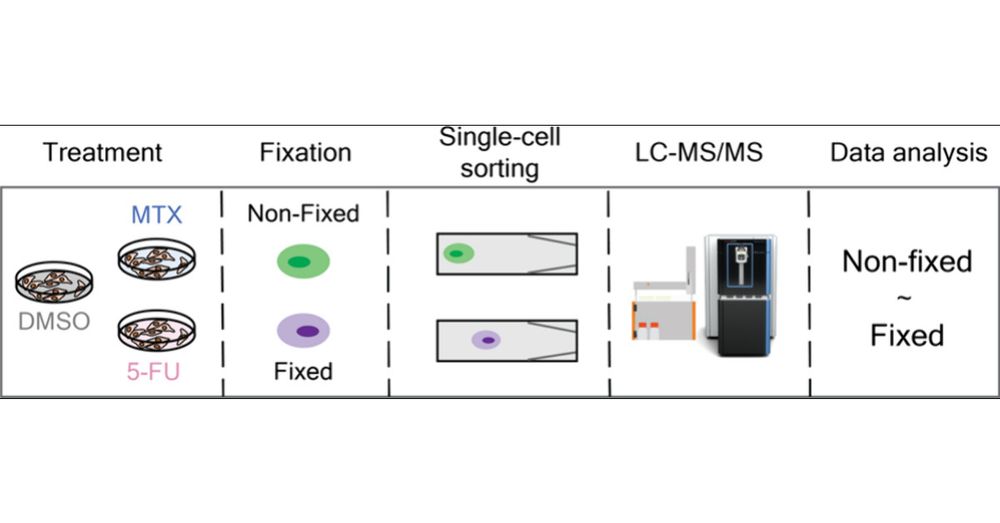
We evaluated how formaldehyde-based fixation preserve proteome state, drug response and cell integrity. Ultimately, cell fixation can facilitate access to #singlecell by enabeling sample shiping and prolonged sorting 📦
pubs.acs.org/doi/10.1021/...

A huge shoutout to my amazing co-authors for their incredible contributions!
Key highlights in replies (1/n)

A huge shoutout to my amazing co-authors for their incredible contributions!
Key highlights in replies (1/n)
23 talks freely accessible, Zoom link on the website.
jnm-2025.mozellosite.com
Jean-Pierre Changeux (allosteric proteins)
en.wikipedia.org/wiki/Jean-Pi...
kicks off at 17:00 GMT
#teammasspec
23 talks freely accessible, Zoom link on the website.
jnm-2025.mozellosite.com
Jean-Pierre Changeux (allosteric proteins)
en.wikipedia.org/wiki/Jean-Pi...
kicks off at 17:00 GMT
#teammasspec
www.nature.com/articles/s41...

www.nature.com/articles/s41...

We describe a label-free single-cell proteomics workflow that enables great depth (>4500 proteins in stem cells) and higher throughput of up to 120 SPD, allowing reliable quantification of key cell markers.
www.nature.com/articles/s41...

We describe a label-free single-cell proteomics workflow that enables great depth (>4500 proteins in stem cells) and higher throughput of up to 120 SPD, allowing reliable quantification of key cell markers.
www.nature.com/articles/s41...
---
#proteomics #prot-paper

---
#proteomics #prot-paper
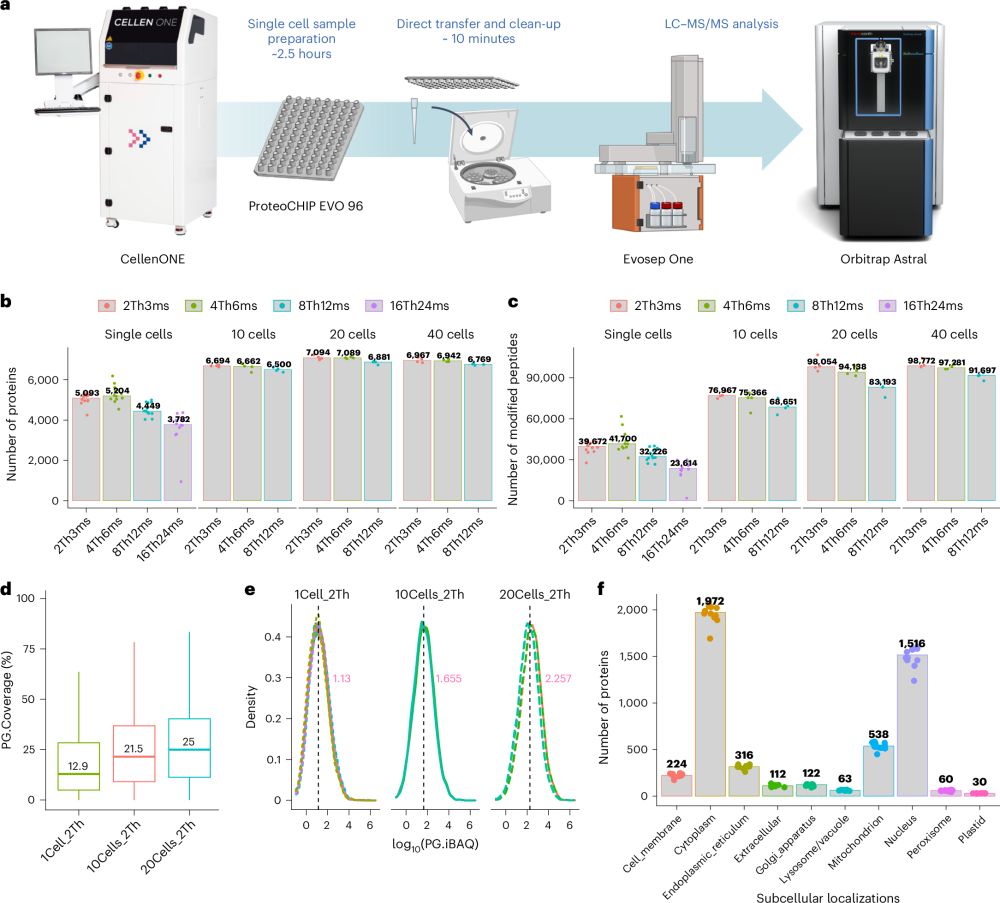

Preprint for our novel implementation of PAC and DIA for REDOX proteomics, with application to cardiovascular diseases♥️🐖🔬. Check it out in @biorxivpreprint.bsky.social
www.biorxiv.org/content/10.1...

Preprint for our novel implementation of PAC and DIA for REDOX proteomics, with application to cardiovascular diseases♥️🐖🔬. Check it out in @biorxivpreprint.bsky.social
www.biorxiv.org/content/10.1...
Pierre Sabatier (@pierresabatier.bsky.social) has. "That's the first time in my career that I felt like I absolutely needed to make it happen."
Learn more about Pierre and read our full discussion over at bit.ly/3VIWkpd


