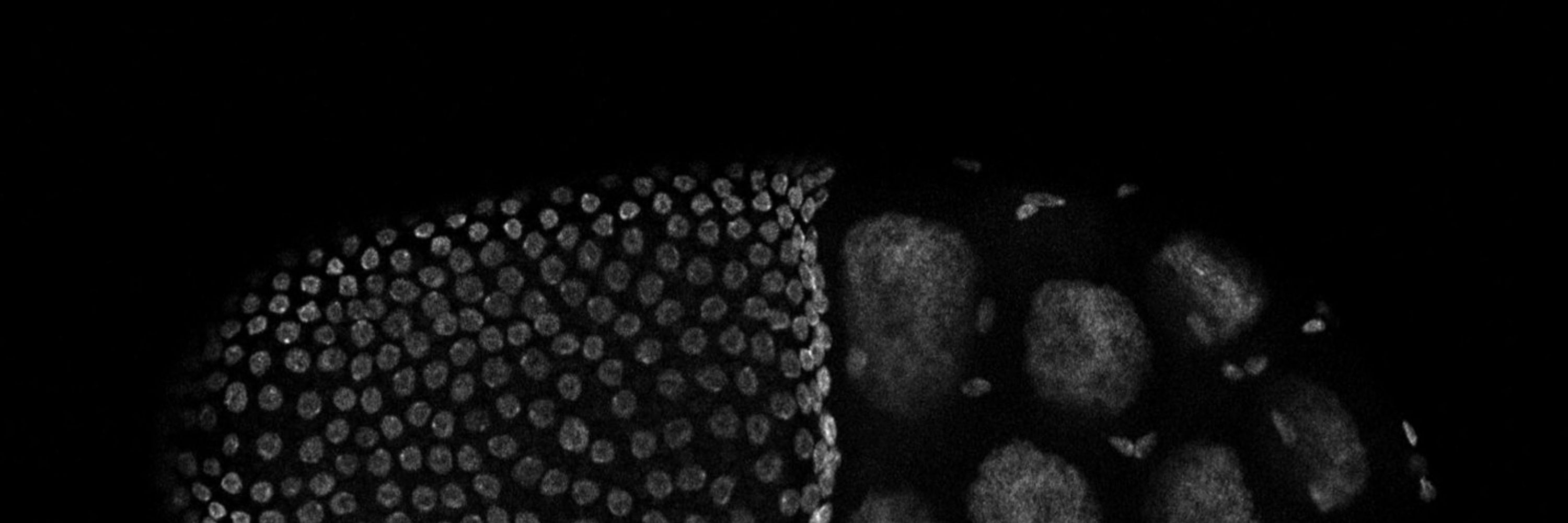
Interested in transcription and RNA export.
http://www.oeaw.ac.at/imba/research/julius-brennecke
TLDR; TFIID behaves differently depending on promoter type. More below:

TLDR; TFIID behaves differently depending on promoter type. More below:
Despite driving ~2/3 of mammalian genes, CpG island (CGI) promoters have remained a puzzle. We identified >50 activators that are exclusively compatible with this promoter class. 🧬
Despite driving ~2/3 of mammalian genes, CpG island (CGI) promoters have remained a puzzle. We identified >50 activators that are exclusively compatible with this promoter class. 🧬
And found a hidden RNA-decay axis from Piwi to the RNA exosome.
And found a hidden RNA-decay axis from Piwi to the RNA exosome.
And found a hidden RNA-decay axis from Piwi to the RNA exosome.
Our study reveals how cohesin guides focused and accurate homology search.
Read more 👉 www.science.org/doi/10.1126/...
Follow along for key insights and updates! 🧵

Our study reveals how cohesin guides focused and accurate homology search.
Read more 👉 www.science.org/doi/10.1126/...
Follow along for key insights and updates! 🧵
on her PhD! Her journey as a joint student (Brennecke/Our Lab) led to key insights into gene silencing. Paper: www.cell.com/molecular-ce.... Journey: www.viennabiocenter.org/about/news/t.... CP.

on her PhD! Her journey as a joint student (Brennecke/Our Lab) led to key insights into gene silencing. Paper: www.cell.com/molecular-ce.... Journey: www.viennabiocenter.org/about/news/t.... CP.
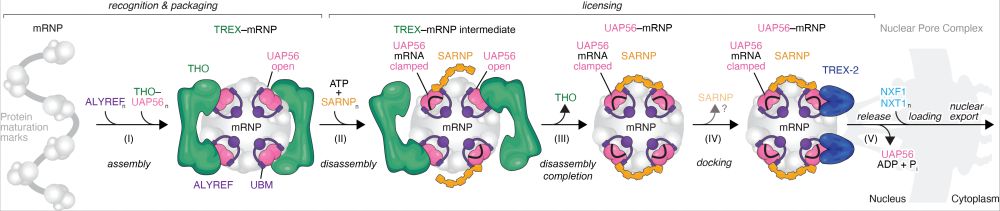
what started as a project on how cells export piRNA precursors, ended up as a tour de force in mRNA export. truly wonderful collaboration with @plaschkalab.bsky.social at the @viennabiocenter.bsky.social
@imbavienna.bsky.social solved a decade-old puzzle, uncovering how the information molecule mRNA travels from the cell’s nucleus to its periphery. More: bit.ly/4nHcvys

what started as a project on how cells export piRNA precursors, ended up as a tour de force in mRNA export. truly wonderful collaboration with @plaschkalab.bsky.social at the @viennabiocenter.bsky.social

🧬We'll be finding funky new RNA biology, mainly by looking at reverse transcriptases (i.e. the Best Enzymes In The World)🧬
annnd: I'm hiring - come join! Especially postdocs and PhD students - please get in touch (NYC is great)

🧬We'll be finding funky new RNA biology, mainly by looking at reverse transcriptases (i.e. the Best Enzymes In The World)🧬
annnd: I'm hiring - come join! Especially postdocs and PhD students - please get in touch (NYC is great)

I am happy to share our new preprint: a chromosome-scale genome assembly for Drosophila OSC cells, one of the key model systems in the piRNA field, especially for nuclear piRNA biology. 🧬🧵 (1/12)

I am happy to share our new preprint: a chromosome-scale genome assembly for Drosophila OSC cells, one of the key model systems in the piRNA field, especially for nuclear piRNA biology. 🧬🧵 (1/12)
We will explore new mechanisms in eukaryotic gene expression, leveraging ‘evolutionary play’ to uncover how regulation, repurposing, and hijacking shape RNA biology.
PhD positions available!
We will explore new mechanisms in eukaryotic gene expression, leveraging ‘evolutionary play’ to uncover how regulation, repurposing, and hijacking shape RNA biology.
PhD positions available!

More info: www.imb.de/students-pos...
More info: www.imb.de/students-pos...
Join us to explore the piRNA pathway with structural and genetic approaches (see 👇👇).
PhD student/postdoc position co-supervised by Clemens Plaschka & myself.
DM or email us if you’d like to know more!
@vbcscitraining.bsky.social @imbavienna.bsky.social @impvienna.bsky.social
But how does piRNA-guided target interaction translate into silencing?
PhD student Júlia Portell Montserrat has an intriguing answer
www.cell.com/molecular-ce...

Join us to explore the piRNA pathway with structural and genetic approaches (see 👇👇).
PhD student/postdoc position co-supervised by Clemens Plaschka & myself.
DM or email us if you’d like to know more!
@vbcscitraining.bsky.social @imbavienna.bsky.social @impvienna.bsky.social
Here we propose a model where a silencing complex, PIWI*, assembles on target RNAs to recruit effectors and shut down transposon activity.
Huge thanks to the Brennecke and Plaschka labs, especially Julius and Clemens, and all co-authors!
But how does piRNA-guided target interaction translate into silencing?
PhD student Júlia Portell Montserrat has an intriguing answer
www.cell.com/molecular-ce...

Here we propose a model where a silencing complex, PIWI*, assembles on target RNAs to recruit effectors and shut down transposon activity.
Huge thanks to the Brennecke and Plaschka labs, especially Julius and Clemens, and all co-authors!
But how does piRNA-guided target interaction translate into silencing?
PhD student Júlia Portell Montserrat has an intriguing answer
www.cell.com/molecular-ce...

But how does piRNA-guided target interaction translate into silencing?
PhD student Júlia Portell Montserrat has an intriguing answer
www.cell.com/molecular-ce...
Julia is the hero of this work, she is currently looking for postdoc labs …
In a collaborative project with the lab of Clemens Plaschka at the IMP, the lab of Julius Brennecke has revealed how PIWI proteins kickstart transposon silencing in reproductive cells.
The findings are published in Molecular Cell: www.sciencedirect.com/science/arti...
Researchers from IMBA and IMP identify the first protein interactions that trigger PIWI–piRNA–mediated transposon silencing, using AlphaFold predictions, genetics, biochemistry and cell biology.
Read more: www.viennabiocenter.org/about/news/t...
Julia is the hero of this work, she is currently looking for postdoc labs …

Researchers at the Vienna BioCenter have solved a 20-year-old mystery in genome biology, revealing how PIWI proteins engage partner molecules to silence “jumping genes” that threaten genetic stability.
Researchers from IMBA and IMP identify the first protein interactions that trigger PIWI–piRNA–mediated transposon silencing, using AlphaFold predictions, genetics, biochemistry and cell biology.
Read more: www.viennabiocenter.org/about/news/t...

Researchers at the Vienna BioCenter have solved a 20-year-old mystery in genome biology, revealing how PIWI proteins engage partner molecules to silence “jumping genes” that threaten genetic stability.
Working with him and Luisa at the IMP was a great and formative experience.
Working with him and Luisa at the IMP was a great and formative experience.
www.biorxiv.org/content/10.1...
www.biorxiv.org/content/10.1...

